The Drosophila peptidoglycan-recognition protein LF interacts with peptidoglycan-recognition protein LC to downregulate the Imd pathway
- PMID: 21372849
- PMCID: PMC3077246
- DOI: 10.1038/embor.2011.19
The Drosophila peptidoglycan-recognition protein LF interacts with peptidoglycan-recognition protein LC to downregulate the Imd pathway
Abstract
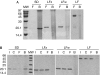
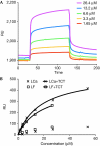
The peptidoglycan (PGN)-recognition protein LF (PGRP-LF) is a specific negative regulator of the immune deficiency (Imd) pathway in Drosophila. We determine the crystal structure of the two PGRP domains constituting the ectodomain of PGRP-LF at 1.72 and 1.94 Å resolution. The structures show that the LFz and LFw domains do not have a PGN-docking groove that is found in other PGRP domains, and they cannot directly interact with PGN, as confirmed by biochemical-binding assays. By using surface plasmon resonance analysis, we show that the PGRP-LF ectodomain interacts with the PGRP-LCx ectodomain in the absence and presence of tracheal cytotoxin. Our results suggest a mechanism for downregulation of the Imd pathway on the basis of the competition between PRGP-LCa and PGRP-LF to bind to PGRP-LCx.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


References
-
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58: 1948–1954 - PubMed
-
- Chang CI, Chelliah Y, Borek D, Mengin-Lecreulx D, Deisenhofer J (2006) Structure of tracheal cytotoxin in complex with a heterodimeric pattern-recognition receptor. Science 311: 1761–1764 - PubMed
-
- Charbonnier S, Zanier K, Masson M, Trave G (2006) Capturing protein–protein complexes at equilibrium: the holdup comparative chromatographic retention assay. Protein Expr Purif 50: 89–101 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

