Activation of multiple proto-oncogenic tyrosine kinases in breast cancer via loss of the PTPN12 phosphatase
- PMID: 21376233
- PMCID: PMC6014607
- DOI: 10.1016/j.cell.2011.02.003
Activation of multiple proto-oncogenic tyrosine kinases in breast cancer via loss of the PTPN12 phosphatase
Abstract
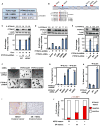
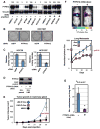
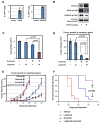
Among breast cancers, triple-negative breast cancer (TNBC) is the most poorly understood and is refractory to current targeted therapies. Using a genetic screen, we identify the PTPN12 tyrosine phosphatase as a tumor suppressor in TNBC. PTPN12 potently suppresses mammary epithelial cell proliferation and transformation. PTPN12 is frequently compromised in human TNBCs, and we identify an upstream tumor-suppressor network that posttranscriptionally controls PTPN12. PTPN12 suppresses transformation by interacting with and inhibiting multiple oncogenic tyrosine kinases, including HER2 and EGFR. The tumorigenic and metastatic potential of PTPN12-deficient TNBC cells is severely impaired upon restoration of PTPN12 function or combined inhibition of PTPN12-regulated tyrosine kinases, suggesting that TNBCs are dependent on the proto-oncogenic tyrosine kinases constrained by PTPN12. Collectively, these data identify PTPN12 as a commonly inactivated tumor suppressor and provide a rationale for combinatorially targeting proto-oncogenic tyrosine kinases in TNBC and other cancers based on their profile of tyrosine-phosphatase activity.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures




Comment in
-
Uncovering a tumor suppressor for triple-negative breast cancers.Cell. 2011 Mar 4;144(5):638-40. doi: 10.1016/j.cell.2011.02.030. Cell. 2011. PMID: 21376226
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

