Choosing the right path: does DNA-PK help make the decision?
- PMID: 21376743
- PMCID: PMC3109507
- DOI: 10.1016/j.mrfmmm.2011.02.010
Choosing the right path: does DNA-PK help make the decision?
Abstract
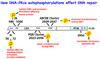
DNA double-strand breaks are extremely harmful lesions that can lead to genomic instability and cell death if not properly repaired. There are at least three pathways that are responsible for repairing DNA double-strand breaks in mammalian cells: non-homologous end joining, homologous recombination and alternative non-homologous end joining. Here we review each of these three pathways with an emphasis on the role of the DNA-dependent protein kinase, a critical component of the non-homologous end joining pathway, in influencing which pathway is ultimately utilized for repair.
2011 Elsevier B.V. All rights reserved.
Figures

References
-
- Mimori T, Hardin JA, Steitz JA. Characterization of the DNA-binding protein antigen Ku recognized by autoantibodies from patients with rheumatic disorders. J. Biol. Chem. 1986;261:2274–2278. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

