Characterisation of the putative effector interaction site of the regulatory HbpR protein from Pseudomonas azelaica by site-directed mutagenesis
- PMID: 21379585
- PMCID: PMC3040749
- DOI: 10.1371/journal.pone.0016539
Characterisation of the putative effector interaction site of the regulatory HbpR protein from Pseudomonas azelaica by site-directed mutagenesis
Abstract
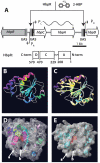
Bacterial transcription activators of the XylR/DmpR subfamily exert their expression control via σ(54)-dependent RNA polymerase upon stimulation by a chemical effector, typically an aromatic compound. Where the chemical effector interacts with the transcription regulator protein to achieve activation is still largely unknown. Here we focus on the HbpR protein from Pseudomonas azelaica, which is a member of the XylR/DmpR subfamily and responds to biaromatic effectors such as 2-hydroxybiphenyl. We use protein structure modeling to predict folding of the effector recognition domain of HbpR and molecular docking to identify the region where 2-hydroxybiphenyl may interact with HbpR. A large number of site-directed HbpR mutants of residues in- and outside the predicted interaction area was created and their potential to induce reporter gene expression in Escherichia coli from the cognate P(C) promoter upon activation with 2-hydroxybiphenyl was studied. Mutant proteins were purified to study their conformation. Critical residues for effector stimulation indeed grouped near the predicted area, some of which are conserved among XylR/DmpR subfamily members in spite of displaying different effector specificities. This suggests that they are important for the process of effector activation, but not necessarily for effector specificity recognition.
Conflict of interest statement
Figures





References
-
- Shingler V. Signal sensing by σ54-dependent regulators: derepression as a control mechanism. Mol Microbiol. 1996;19:409–416. - PubMed
-
- Ramos JL, Marques S, Timmis KN. Transcriptional control of the Pseudomonas TOL plasmid catabolic operons is achieved through an interplay of host factors and plasmid- encoded regulators. Annu Rev Microbiol. 1997;51:341–373. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

