Specific binding of adamantane drugs and direction of their polar amines in the pore of the influenza M2 transmembrane domain in lipid bilayers and dodecylphosphocholine micelles determined by NMR spectroscopy
- PMID: 21381693
- PMCID: PMC3078525
- DOI: 10.1021/ja102581n
Specific binding of adamantane drugs and direction of their polar amines in the pore of the influenza M2 transmembrane domain in lipid bilayers and dodecylphosphocholine micelles determined by NMR spectroscopy
Abstract
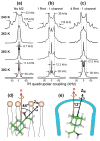
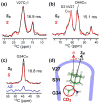
The transmembrane domain of the influenza M2 protein (M2TM) forms a tetrameric proton channel important for the virus lifecycle. The proton-channel activity is inhibited by amine-containing adamantyl drugs amantadine and rimantadine, which have been shown to bind specifically to the pore of M2TM near Ser31. However, whether the polar amine points to the N- or C-terminus of the channel has not yet been determined. Elucidating the polar group direction will shed light on the mechanism by which drug binding inhibits this proton channel and will facilitate rational design of new inhibitors. In this study, we determine the polar amine direction using M2TM reconstituted in lipid bilayers as well as dodecylphosphocholine (DPC) micelles. (13)C-(2)H rotational-echo double-resonance NMR experiments of (13)C-labeled M2TM and methyl-deuterated rimantadine in lipid bilayers showed that the polar amine pointed to the C-terminus of the channel, with the methyl group close to Gly34. Solution NMR experiments of M2TM in DPC micelles indicate that drug binding causes significant chemical shift perturbations of the protein that are very similar to those seen for M2TM and M2(18-60) bound to lipid bilayers. Specific (2)H-labeling of the drugs permitted the assignment of drug-protein cross peaks, which indicate that amantadine and rimantadine bind to the pore in the same fashion as for bilayer-bound M2TM. These results strongly suggest that adamantyl inhibition of M2TM is achieved not only by direct physical occlusion of the channel, but also by perturbing the equilibrium constant of the proton-sensing residue His37. The reproduction of the pharmacologically relevant specific pore-binding site in DPC micelles, which was not observed with a different detergent, DHPC, underscores the significant influence of the detergent environment on the functional structure of this membrane protein.
Figures



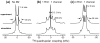

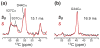

References
-
- Pinto LH, Lamb RA. J Biol Chem. 2006;281:8997–9000. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

