Evolutionary transients in the rice transcriptome
- PMID: 21382590
- PMCID: PMC5054128
- DOI: 10.1016/S1672-0229(10)60023-X
Evolutionary transients in the rice transcriptome
Abstract
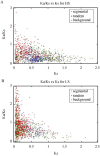
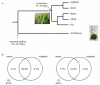
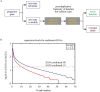
In the canonical version of evolution by gene duplication, one copy is kept unaltered while the other is free to evolve. This process of evolutionary experimentation can persist for millions of years. Since it is so short lived in comparison to the lifetime of the core genes that make up the majority of most genomes, a substantial fraction of the genome and the transcriptome may-in principle-be attributable to what we will refer to as "evolutionary transients", referring here to both the process and the genes that have gone or are undergoing this process. Using the rice gene set as a test case, we argue that this phenomenon goes a long way towards explaining why there are so many more rice genes than Arabidopsis genes, and why most excess rice genes show low similarity to eudicots.
Copyright © 2010 Beijing Genomics Institute. Published by Elsevier Ltd. All rights reserved.
Figures


References
-
- Lynch M., Conery J.S. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. - PubMed
-
- International Rice Genome Sequencing Project The map-based sequence of the rice genome. Nature. 2005;436:793–800. - PubMed
-
- The Arabidopsis Genome Initiative Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. - PubMed
-
- Yu J. A draft sequence of the rice genome (Oryza sativa L. ssp. indica) Science. 2002;296:79–92. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

