Defining the earliest step of cardiovascular progenitor specification during embryonic stem cell differentiation
- PMID: 21383076
- PMCID: PMC3051813
- DOI: 10.1083/jcb.201007063
Defining the earliest step of cardiovascular progenitor specification during embryonic stem cell differentiation
Abstract
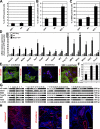
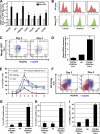
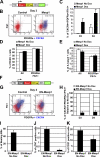
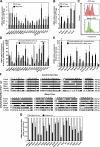
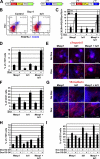
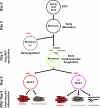
During embryonic development and embryonic stem cell (ESC) differentiation, the different cell lineages of the mature heart arise from two types of multipotent cardiovascular progenitors (MCPs), the first and second heart fields. A key question is whether these two MCP populations arise from differentiation of a common progenitor. In this paper, we engineered Mesp1-green fluorescent protein (GFP) ESCs to isolate early MCPs during ESC differentiation. Mesp1-GFP cells are strongly enriched for MCPs, presenting the ability to differentiate into multiple cardiovascular lineages from both heart fields in vitro and in vivo. Transcriptional profiling of Mesp1-GFP cells uncovered cell surface markers expressed by MCPs allowing their prospective isolation. Mesp1 is required for MCP specification and the expression of key cardiovascular transcription factors. Isl1 is expressed in a subset of early Mesp1-expressing cells independently of Mesp1 and acts together with Mesp1 to promote cardiovascular differentiation. Our study identifies the early MCPs residing at the top of the cellular hierarchy of cardiovascular lineages during ESC differentiation.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

