Recurrence, submicroscopic complexity, and potential clinical relevance of copy gains detected by array CGH that are shown to be unbalanced insertions by FISH
- PMID: 21383316
- PMCID: PMC3065701
- DOI: 10.1101/gr.114579.110
Recurrence, submicroscopic complexity, and potential clinical relevance of copy gains detected by array CGH that are shown to be unbalanced insertions by FISH
Abstract
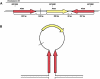
Insertions occur when a segment of one chromosome is translocated and inserted into a new region of the same chromosome or a non-homologous chromosome. We report 71 cases with unbalanced insertions identified using array CGH and FISH in 4909 cases referred to our laboratory for array CGH and found to have copy-number abnormalities. Although the majority of insertions were non-recurrent, several recurrent unbalanced insertions were detected, including three der(Y)ins(Y;18)(q?11.2;p11.32p11.32)pat inherited from parents carrying an unbalanced insertion. The clinical significance of these recurrent rearrangements is unclear, although the small size, limited gene content, and inheritance pattern of each suggests that the phenotypic consequences may be benign. Cryptic, submicroscopic duplications were observed at or near the insertion sites in two patients, further confounding the clinical interpretation of these insertions. Using FISH, linear amplification, and array CGH, we identified a 126-kb duplicated region from 19p13.3 inserted into MECP2 at Xq28 in a patient with symptoms of Rett syndrome. Our results demonstrate that although the interpretation of most non-recurrent insertions is unclear without high-resolution insertion site characterization, the potential for an otherwise benign duplication to result in a clinically relevant outcome through the disruption of a gene necessitates the use of FISH to determine whether copy-number gains detected by array CGH represent tandem duplications or unbalanced insertions. Further follow-up testing using techniques such as linear amplification or sequencing should be used to determine gene involvement at the insertion site after FISH has identified the presence of an insertion.
Figures




References
-
- Abuelo DN, Barsel-Bowers G, Richardson A 1988. Insertional translocations: Report of two new families and review of the literature. Am J Med Genet 31: 319–329 - PubMed
-
- Ballif BC, Wakui K, Gajecka M, Shaffer LG 2004. Translocation breakpoint mapping and sequence analysis in three monosomy 1p36 subjects with der(1)t(1;1)(p36;q44) suggest mechanisms for telomere capture in stabilizing de novo terminal rearrangements. Hum Genet 114: 198–206 - PubMed
-
- Ballif BC, Theisen A, Coppinger J, Gowans GC, Hersh JH, Madan-Khetarpal S, Schmidt KR, Tervo R, Escobar LF, Friedrich CA, et al. 2008a. Expanding the clinical phenotype of the 3q29 microdeletion syndrome and characterization of the reciprocal microduplication. Mol Cytogenet 1: 8 doi: 10.1186/1755-8166-1-8 - PMC - PubMed
-
- Ballif BC, Theisen A, McDonald-McGinn DM, Zackai EH, Hersh JH, Bejjani BA, Shaffer LG 2008b. Identification of a previously unrecognized microdeletion syndrome of 16q11.2q12.2. Clin Genet 74: 469–475 - PubMed
-
- Baptista J, Prigmore E, Gribble SM, Jacobs PA, Carter NP, Crolla JA 2005. Molecular cytogenetic analyses of breakpoints in apparently balanced reciprocal translocations carried by phenotypically normal individuals. Eur J Hum Genet 13: 1205–1212 - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
