Genome-wide association study identifies Loci for body composition and structural soundness traits in pigs
- PMID: 21383979
- PMCID: PMC3044704
- DOI: 10.1371/journal.pone.0014726
Genome-wide association study identifies Loci for body composition and structural soundness traits in pigs
Abstract
Background: The recent completion of the swine genome sequencing project and development of a high density porcine SNP array has made genome-wide association (GWA) studies feasible in pigs.
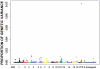
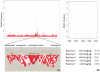
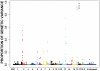
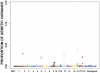
Methodology/principal findings: Using Illumina's PorcineSNP60 BeadChip, we performed a pilot GWA study in 820 commercial female pigs phenotyped for backfat, loin muscle area, body conformation in addition to feet and leg (FL) structural soundness traits. A total of 51,385 SNPs were jointly fitted using Bayesian techniques as random effects in a mixture model that assumed a known large proportion (99.5%) of SNPs had zero effect. SNP annotations were implemented through the Sus scrofa Build 9 available from pig Ensembl. We discovered a number of candidate chromosomal regions, and some of them corresponded to QTL regions previously reported. We not only have identified some well-known candidate genes for the traits of interest, such as MC4R (for backfat) and IGF2 (for loin muscle area), but also obtained novel promising genes, including CHCHD3 (for backfat), BMP2 (for loin muscle area, body size and several FL structure traits), and some HOXA family genes (for overall leg action). The candidate regions responsible for body conformation and FL structure soundness did not overlap greatly which implied that these traits were controlled by different genes. Functional clustering analyses classified the genes into categories related to bone and cartilage development, muscle growth and development or the insulin pathway suggesting the traits are regulated by common pathways or gene networks that exert roles at different spatial and temporal stages.
Conclusions/significance: This study is one of the earliest GWA reports on important quantitative traits in pigs, and the findings will contribute to the further biological function analysis of the identified candidate genes and potential utilization of them in marker assisted selection.
Conflict of interest statement
Figures
References
-
- Albarella U, Dobney K, Ervynck A, Rowley-Conwy P. New York: Oxford University Press Inc; 2007. Pigs and Human: 10,000 years of interaction.11
-
- Andersson L, Georges M. Domestic-animal genomics: Deciphering the genetics of complex traits. Nat Rev Genet. 2004;5:202–212. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

