GST pi modulates JNK activity through a direct interaction with JNK substrate, ATF2
- PMID: 21384452
- PMCID: PMC3125868
- DOI: 10.1002/pro.609
GST pi modulates JNK activity through a direct interaction with JNK substrate, ATF2
Abstract
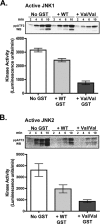
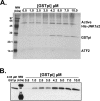
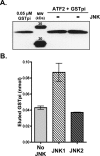
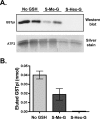
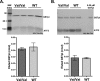
Human GSTpi, an important detoxification enzyme, has been shown to modulate the activity of JNKs by inhibiting apoptosis and by causing cell proliferation and tumor growth. In this work, we describe a detailed analysis of the interaction in vitro between GSTpi and JNK isoforms (both in their inactive and active, phosphorylated forms). The ability of active JNK1 or JNK2 to phosphorylate their substrate, ATF2, is inhibited by two naturally occurring GSTpi haplotypes (Ile105/Ala114, WT or haplotype A, and Val105/Val114, haplotype C). Haplotype C of GSTpi is a more potent inhibitor of JNK activity than haplotype A, yielding 75-80% and 25-45% inhibition, respectively. We show that GSTpi is not a substrate of JNK, as was earlier suggested by others. Through binding studies, we demonstrate that the interaction between GSTpi and phosphorylated, active JNKs is isoform specific, with JNK1 being the preferred isoform. In contrast, GSTpi does not interact with unphosphorylated, inactive JNKs unless a JNK substrate, ATF2, is present. We also demonstrate, for the first time, a direct interaction: between GSTpi and ATF2. GSTpi binds with similar affinity to active JNK + ATF2 and to ATF2 alone. Direct binding experiments between ATF2 and GSTpi, either alone or in the presence of glutathione analogs or phosphorylated ATF2, indicate that the xenobiotic portion of the GSTpi active site and the JNK binding domain of ATF2 are involved in this interaction. Competition between GSTpi and active JNK for the substrate ATF2 may be responsible for the inhibition of JNK catalysis by GSTpi.
Copyright © 2011 The Protein Society.
Figures



References
-
- Holley SL, Fryer AA, Haycock JW, Grubb SE, Strange RC, Hoban PR. Differential effects of glutathione S-transferase pi (GSTP1) haplotypes on cell proliferation and apoptosis. Carcinogenesis. 2007;28:2268–2273. - PubMed
-
- Henderson C, Wolf CR. Disruption of the glutathione transferase Pi class genes. In: Sies H, Packer L, editors. Hoboken, NJ: Methods in enzymology, Vol.401 Elsevier Academic Press; 2005. pp. 116–137. - PubMed
-
- Watson MA, Stewart RK, Smith GB, Massey TE, Bell DA. Human glutathione S-transferase P1 polymorphisms: relationship to lung tissue enzyme activity and population frequency distribution. Carcinogenesis. 1998;19:275–280. - PubMed
-
- Ralat LA, Manevich Y, Fisher AB, Colman RF. Direct evidence for the formation of a complex between 1-cysteine peroxiredoxin and glutathione S-transferase pi with activity changes in both enzymes. Biochemistry. 2006;45:360–372. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

