Altered architecture of substrate binding region defines the unique specificity of UDP-GalNAc 4-epimerases
- PMID: 21384454
- PMCID: PMC3125870
- DOI: 10.1002/pro.611
Altered architecture of substrate binding region defines the unique specificity of UDP-GalNAc 4-epimerases
Abstract
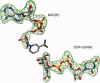
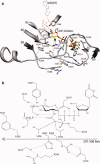
UDP-hexose 4-epimerases play a pivotal role in lipopolysaccharide (LPS) biosynthesis and Leloir pathway. These epimerases are classified into three groups based on whether they recognize nonacetylated UDP-hexoses (Group 1), both N-acetylated and nonacetylated UDP-hexoses (Group 2) or only N-acetylated UDP-hexoses (Group 3). Although the catalysis has been investigated extensively, yet a definitive model rationalizing the substrate specificity of all the three groups on a common platform is largely lacking. In this work, we present the crystal structure of WbgU, a novel UDP-hexose 4-epimerase that belongs to the Group 3. WbgU is involved in biosynthetic pathway of the unusual glycan 2-deoxy-L-altruronic acid that is found in the LPS of the pathogen Pleisomonas shigelloides. A model that defines its substrate specificity is proposed on the basis of the active site architecture. Representatives from all the three groups are then compared to rationalize their substrate specificity. This investigation reveals that the Group 3 active site architecture is markedly different from the "conserved scaffold" of the Group 1 and the Group 2 epimerases and highlights the interactions potentially responsible for the origin of specificity of the Group 3 epimerases toward N-acetylated hexoses. This study provides a platform for further engineering of the UDP-hexose 4-epimerases, leads to a deeper understanding of the LPS biosynthesis and carbohydrate recognition by proteins. It may also have implications in development of novel antibiotics and more economic synthesis of UDP-GalNAc and downstream products such as carbohydrate based vaccines.
Copyright © 2011 The Protein Society.
Figures
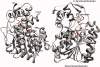

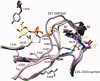
References
-
- Bauer AJ, Rayment I, Frey PA, Holden HM. The molecular structure of UDP-galactose 4-epimerase from Escherichia coli determined at 2.5 A resolution. Proteins. 1992;12:372–381. - PubMed
-
- Kowal P, Wang P. New UDP-GlcNAc C4 epimerase involved in the biosynthesis of 2-acetamino-2-deoxy-l-altruronic acid in the O-antigen repeating units of Plesiomonas shigelloides O17. Biochemistry. 2002;41:15410–15414. - PubMed
-
- Ishiyama N, Creuzenet C, Lam JS, Berghuis AM. Crystal structure of WbpP, a genuine UDP-N-acetylglucosamine 4-epimerase from Pseudomonas aeruginosa: substrate specificity in udp-hexose 4-epimerases. J Biol Chem. 2004;279:22635–22642. - PubMed
-
- Holden HM, Rayment I, Thoden JB. Structure and function of enzymes of the Leloir pathway for galactose metabolism. J Biol Chem. 2003;278:43885–43888. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

