Requirement of TFIIH kinase subunit Mat1 for RNA Pol II C-terminal domain Ser5 phosphorylation, transcription and mRNA turnover
- PMID: 21385826
- PMCID: PMC3130277
- DOI: 10.1093/nar/gkr107
Requirement of TFIIH kinase subunit Mat1 for RNA Pol II C-terminal domain Ser5 phosphorylation, transcription and mRNA turnover
Abstract
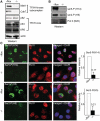
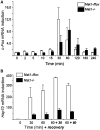
The relevance of serine 5 phosphorylation of RNA polymerase II carboxy-terminal domain during initiation has been difficult to determine in mammalian cells as no general in vivo Ser5 kinase has been identified. Here, we demonstrate that deletion of the TFIIH kinase subunit Mat1 in mouse fibroblasts leads to dramatically reduced Pol II Ser5 phosphorylation. This is associated with defective capping and reduced Ser2 phosphorylation, decreased Pol II progression into elongation and severely attenuated transcription detected through analysis of nascent mRNAs, establishing a general requirement for mammalian Mat1 in transcription. Surprisingly, the general defect in Pol II transcription in Mat1(-/-) fibroblasts is not reflected in the majority of steady-state mRNAs. This indicates widespread stabilization of mRNAs and points to the existence of a regulatory mechanism to stabilize mRNAs following transcriptional attenuation, thus revealing a potential caveat in similar studies limited to analysis of steady-state mRNAs.
Figures

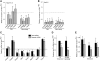
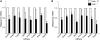
References
-
- Saunders A, Core LJ, Lis JT. Breaking barriers to transcription elongation. Nat. Rev. 2006;7:557–567. - PubMed
-
- Tirode F, Busso D, Coin F, Egly JM. Reconstitution of the transcription factor TFIIH: assignment of functions for the three enzymatic subunits, XPB, XPD, and cdk7. Mol. Cell. 1999;3:87–95. - PubMed
-
- Coin F, Oksenych V, Mocquet V, Groh S, Blattner C, Egly JM. Nucleotide excision repair driven by the dissociation of CAK from TFIIH. Mol. Cell. 2008;31:9–20. - PubMed
-
- Roy R, Adamczewski JP, Seroz T, Vermeulen W, Tassan JP, Schaeffer L, Nigg EA, Hoeijmakers JH, Egly JM. The MO15 cell cycle kinase is associated with the TFIIH transcription- DNA repair factor. Cell. 1994;79:1093–1101. - PubMed
-
- Serizawa H, Makela TP, Conaway JW, Conaway RC, Weinberg RA, Young RA. Association of Cdk-activating kinase subunits with transcription factor TFIIH. Nature. 1995;374:280–282. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

