Maternal diet and aging alter the epigenetic control of a promoter-enhancer interaction at the Hnf4a gene in rat pancreatic islets
- PMID: 21385945
- PMCID: PMC3069181
- DOI: 10.1073/pnas.1019007108
Maternal diet and aging alter the epigenetic control of a promoter-enhancer interaction at the Hnf4a gene in rat pancreatic islets
Abstract
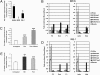
Environmental factors interact with the genome throughout life to determine gene expression and, consequently, tissue function and disease risk. One such factor that is known to play an important role in determining long-term metabolic health is diet during critical periods of development. Epigenetic regulation of gene expression has been implicated in mediating these programming effects of early diet. The precise epigenetic mechanisms that underlie these effects remain largely unknown. Here, we show that the transcription factor Hnf4a, which has been implicated in the etiology of type 2 diabetes (T2D), is epigenetically regulated by maternal diet and aging in rat islets. Transcriptional activity of Hnf4a in islets is restricted to the distal P2 promoter through its open chromatin configuration and an islet-specific interaction between the P2 promoter and a downstream enhancer. Exposure to suboptimal nutrition during early development leads to epigenetic silencing at the enhancer region, which weakens the P2 promoter-enhancer interaction and results in a permanent reduction in Hnf4a expression. Aging leads to progressive epigenetic silencing of the entire Hnf4a locus in islets, an effect that is more pronounced in rats exposed to a poor maternal diet. Our findings provide evidence for environmentally induced epigenetic changes at the Hnf4a enhancer that alter its interaction with the P2 promoter, and consequently determine T2D risk. We therefore propose that environmentally induced changes in promoter-enhancer interactions represent a fundamental epigenetic mechanism by which nutrition and aging can influence long-term health.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
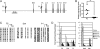


References
-
- Jaenisch R, Bird A. Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–254. - PubMed
-
- Sandovici I, Smith NH, Ozanne SE, Constância M. In: Epigenetics. Tost J, editor. Norfolk, UK: Caister Academic; 2008. pp. 343–370.
-
- Burdge GC, Lillycrop KA. Nutrition, epigenetics, and developmental plasticity: Implications for understanding human disease. Annu Rev Nutr. 2010;30:315–339. - PubMed
-
- Ozanne SE, Constância M. Mechanisms of disease: The developmental origins of disease and the role of the epigenotype. Nat Clin Pract Endocrinol Metab. 2007;3:539–546. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BB/D01235X/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- BB/E00797X/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/D01235X/2/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- FS/09/029/27902/BHF_/British Heart Foundation/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

