Phosphorylation of the Nicotiana benthamiana WRKY8 transcription factor by MAPK functions in the defense response
- PMID: 21386030
- PMCID: PMC3082260
- DOI: 10.1105/tpc.110.081794
Phosphorylation of the Nicotiana benthamiana WRKY8 transcription factor by MAPK functions in the defense response
Abstract
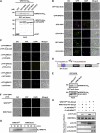
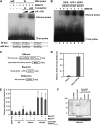
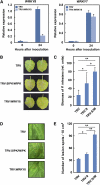
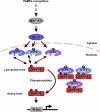
Mitogen-activated protein kinase (MAPK) cascades have pivotal roles in plant innate immunity. However, downstream signaling of plant defense-related MAPKs is not well understood. Here, we provide evidence that the Nicotiana benthamiana WRKY8 transcription factor is a physiological substrate of SIPK, NTF4, and WIPK. Clustered Pro-directed Ser residues (SP cluster), which are conserved in group I WRKY proteins, in the N-terminal region of WRKY8 were phosphorylated by these MAPKs in vitro. Antiphosphopeptide antibodies indicated that Ser residues in the SP cluster of WRKY8 are phosphorylated by SIPK, NTF4, and WIPK in vivo. The interaction of WRKY8 with MAPKs depended on its D domain, which is a MAPK-interacting motif, and this interaction was required for effective phosphorylation of WRKY8 in plants. Phosphorylation of WRKY8 increased its DNA binding activity to the cognate W-box sequence. The phospho-mimicking mutant of WRKY8 showed higher transactivation activity, and its ectopic expression induced defense-related genes, such as 3-hydroxy-3-methylglutaryl CoA reductase 2 and NADP-malic enzyme. By contrast, silencing of WRKY8 decreased the expression of defense-related genes and increased disease susceptibility to the pathogens Phytophthora infestans and Colletotrichum orbiculare. Thus, MAPK-mediated phosphorylation of WRKY8 has an important role in the defense response through activation of downstream genes.
Figures


References
-
- Asai S., Mase K., Yoshioka H. (2010). A key enzyme for flavin synthesis is required for nitric oxide and reactive oxygen species production in disease resistance. Plant J. 62: 911–924 - PubMed
-
- Asai T., Tena G., Plotnikova J., Willmann M.R., Chiu W.L., Gomez-Gomez L., Boller T., Ausubel F.M., Sheen J. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

