Overexpression of Arabidopsis ECERIFERUM1 promotes wax very-long-chain alkane biosynthesis and influences plant response to biotic and abiotic stresses
- PMID: 21386033
- PMCID: PMC3091054
- DOI: 10.1104/pp.111.172320
Overexpression of Arabidopsis ECERIFERUM1 promotes wax very-long-chain alkane biosynthesis and influences plant response to biotic and abiotic stresses
Abstract
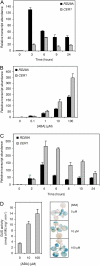
Land plant aerial organs are covered by a hydrophobic layer called the cuticle that serves as a waterproof barrier protecting plants against desiccation, ultraviolet radiation, and pathogens. Cuticle consists of a cutin matrix as well as cuticular waxes in which very-long-chain (VLC) alkanes are the major components, representing up to 70% of the total wax content in Arabidopsis (Arabidopsis thaliana) leaves. However, despite its major involvement in cuticle formation, the alkane-forming pathway is still largely unknown. To address this deficiency, we report here the characterization of the Arabidopsis ECERIFERUM1 (CER1) gene predicted to encode an enzyme involved in alkane biosynthesis. Analysis of CER1 expression showed that CER1 is specifically expressed in the epidermis of aerial organs and coexpressed with other genes of the alkane-forming pathway. Modification of CER1 expression in transgenic plants specifically affects VLC alkane biosynthesis: waxes of TDNA insertional mutant alleles are devoid of VLC alkanes and derivatives, whereas CER1 overexpression dramatically increases the production of the odd-carbon-numbered alkanes together with a substantial accumulation of iso-branched alkanes. We also showed that CER1 expression is induced by osmotic stresses and regulated by abscisic acid. Furthermore, CER1-overexpressing plants showed reduced cuticle permeability together with reduced soil water deficit susceptibility. However, CER1 overexpression increased susceptibility to bacterial and fungal pathogens. Taken together, these results demonstrate that CER1 controls alkane biosynthesis and is highly linked to responses to biotic and abiotic stresses.
Figures





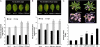
References
-
- Aguirrezabal L, Bouchier-Combaud S, Radziejwoski A, Dauzat M, Cookson SJ, Granier C. (2006) Plasticity to soil water deficit in Arabidopsis thaliana: dissection of leaf development into underlying growth dynamic and cellular variables reveals invisible phenotypes. Plant Cell Environ 29: 2216–2227 - PubMed
-
- Ariizumi T, Hatakeyama K, Hinata K, Sato S, Kato T, Tabata S, Toriyama K. (2003) A novel male-sterile mutant of Arabidopsis thaliana, faceless pollen-1, produces pollen with a smooth surface and an acetolysis-sensitive exine. Plant Mol Biol 53: 107–116 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

