The expanded human disease network combining protein-protein interaction information
- PMID: 21386875
- PMCID: PMC3137500
- DOI: 10.1038/ejhg.2011.30
The expanded human disease network combining protein-protein interaction information
Abstract
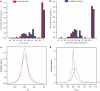
The human disease network (HDN) has become a powerful tool for revealing disease-disease associations. Some studies have shown that genes that share similar or same disease phenotypes tend to encode proteins that interact with each other. Therefore, protein-protein interactions (PPIs) may help us to further understand the relationships between diseases with overlapping clinical phenotypes. In this study, we constructed the expanded HDN (eHDN) by combining disease gene information with PPI information, and analyzed its topological features and functional properties. We found that the network is hierarchical and, most diseases are connected to only a few diseases, whereas a small part of diseases are linked to many different diseases. Diseases in a specific disease class tend to cluster together, and genes associated with the same disease are functionally related. Comparing the eHDN with the original HDN (oHDN, constructed using disease gene information) revealed high consistency over all topological and functional properties. This, to some extent, indicates that our eHDN is reliable. In the eHDN, we found some new associations among diseases resulting from the shared genes interacting with disease genes. The new eHDN will provide a valuable reference for clinicians and medical researchers.
Figures

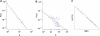
References
-
- Barabasi AL, Oltvai ZN. Network biology: understanding the cell's functional organization. Nat Rev Genet. 2004;5:101–113. - PubMed
-
- Ge H, Walhout AJ, Vidal M. Integrating ‘omic' information: a bridge between genomics and systems biology. Trends Genet. 2003;19:551–560. - PubMed
-
- Yook SH, Oltvai ZN, Barabasi AL. Functional and topological characterization of protein interaction networks. Proteomics. 2004;4:928–942. - PubMed
-
- Shen-Orr SS, Milo R, Mangan S, Alon U. Network motifs in the transcriptional regulation network of Escherichia coli. Nat Genet. 2002;31:64–68. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

