Genomic profiling of advanced-stage oral cancers reveals chromosome 11q alterations as markers of poor clinical outcome
- PMID: 21386901
- PMCID: PMC3046132
- DOI: 10.1371/journal.pone.0017250
Genomic profiling of advanced-stage oral cancers reveals chromosome 11q alterations as markers of poor clinical outcome
Abstract
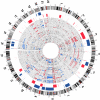
Identifying oral cancer lesions associated with high risk of relapse and predicting clinical outcome remain challenging questions in clinical practice. Genomic alterations may add prognostic information and indicate biological aggressiveness thereby emphasizing the need for genome-wide profiling of oral cancers. High-resolution array comparative genomic hybridization was performed to delineate the genomic alterations in clinically annotated primary gingivo-buccal complex and tongue cancers (n = 60). The specific genomic alterations so identified were evaluated for their potential clinical relevance. Copy-number changes were observed on chromosomal arms with most frequent gains on 3q (60%), 5p (50%), 7p (50%), 8q (73%), 11q13 (47%), 14q11.2 (47%), and 19p13.3 (58%) and losses on 3p14.2 (55%) and 8p (83%). Univariate statistical analysis with correction for multiple testing revealed chromosomal gain of region 11q22.1-q22.2 and losses of 17p13.3 and 11q23-q25 to be associated with loco-regional recurrence (P = 0.004, P = 0.003, and P = 0.0003) and shorter survival (P = 0.009, P = 0.003, and P 0.0001) respectively. The gain of 11q22 and loss of 11q23-q25 were validated by interphase fluorescent in situ hybridization (I-FISH). This study identifies a tractable number of genomic alterations with few underlying genes that may potentially be utilized as biological markers for prognosis and treatment decisions in oral cancers.
Conflict of interest statement
Figures

References
-
- Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. - PubMed
-
- Lippman SM, Hong WK. Molecular markers of the risk of oral cancer. N Engl J Med. 2001;344:1323–1326. - PubMed
-
- Akervall J. Genomic screening of head and neck cancer and its implications for therapy planning. Eur Arch Otorhinolaryngol. 2006;263:297–304. - PubMed
-
- Solinas-Toldo S, Lampel S, Stilgenbauer S, Nickolenko J, Benner A, et al. Matrix-based comparative genomic hybridization: biochips to screen for genomic imbalances. Genes Chromosomes Cancer. 1997;20:399–407. - PubMed
-
- Kallioniemi A. CGH microarrays and cancer. Curr Opin Biotechnol. 2008;19:36–40. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

