New implications on genomic adaptation derived from the Helicobacter pylori genome comparison
- PMID: 21387011
- PMCID: PMC3046158
- DOI: 10.1371/journal.pone.0017300
New implications on genomic adaptation derived from the Helicobacter pylori genome comparison
Erratum in
- PLoS One. 2011;6(3). doi: 10.1371/annotation/2f2953da-1918-4068-a730-46660d43e6b5 doi: 10.1371/annotation/2f2953da-1918-4068-a730-46660d43e6b5
- PLoS One. 2011;6(3). doi: 10.1371/annotation/d8fae120-5f86-4df7-8e53-46afa9c01be0 doi: 10.1371/annotation/d8fae120-5f86-4df7-8e53-46afa9c01be0
Abstract
Background: Helicobacter pylori has a reduced genome and lives in a tough environment for long-term persistence. It evolved with its particular characteristics for biological adaptation. Because several H. pylori genome sequences are available, comparative analysis could help to better understand genomic adaptation of this particular bacterium.
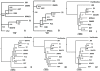
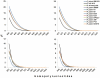
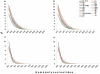
Principal findings: We analyzed nine H. pylori genomes with emphasis on microevolution from a different perspective. Inversion was an important factor to shape the genome structure. Illegitimate recombination not only led to genomic inversion but also inverted fragment duplication, both of which contributed to the creation of new genes and gene family, and further, homological recombination contributed to events of inversion. Based on the information of genomic rearrangement, the first genome scaffold structure of H. pylori last common ancestor was produced. The core genome consists of 1186 genes, of which 22 genes could particularly adapt to human stomach niche. H. pylori contains high proportion of pseudogenes whose genesis was principally caused by homopolynucleotide (HPN) mutations. Such mutations are reversible and facilitate the control of gene expression through the change of DNA structure. The reversible mutations and a quasi-panmictic feature could allow such genes or gene fragments frequently transferred within or between populations. Hence, pseudogenes could be a reservoir of adaptation materials and the HPN mutations could be favorable to H. pylori adaptation, leading to HPN accumulation on the genomes, which corresponds to a special feature of Helicobacter species: extremely high HPN composition of genome.
Conclusion: Our research demonstrated that both genome content and structure of H. pylori have been highly adapted to its particular life style.
Conflict of interest statement
Figures







References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

