Insights into MHC class I antigen processing gained from large-scale analysis of class I ligands
- PMID: 21387142
- PMCID: PMC11114492
- DOI: 10.1007/s00018-011-0659-9
Insights into MHC class I antigen processing gained from large-scale analysis of class I ligands
Abstract
Short peptides derived from intracellular proteins and presented on MHC class I molecules on the cell surface serve as a showcase for the immune system to detect pathogenic or malignant alterations inside the cell, and the sequencing and analysis of the presented peptide pool has received considerable attention over the last two decades. In this review, we give a comprehensive presentation of the methods employed for the large-scale qualitative and quantitative analysis of the MHC class I ligandome. Furthermore, we focus on insights gained into the underlying processing pathway, especially involving the roles of the proteasome, the TAP complex, and the peptide specificities and motifs of MHC molecules. The identification of post-translational modifications in MHC ligands and their implications for processing are also considered. Finally, we review the correlations of the ligandome to the proteome and the transcriptome.
Figures
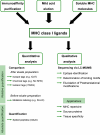

References
-
- Rammensee HG, Falk K, Rötzschke O. Peptides naturally presented by MHC class I molecules. Annu Rev Immunol. 1993;11:213–244. - PubMed
-
- Stevanović S, Schild H. Quantitative aspects of T cell activation—peptide generation and editing by MHC class I molecules. Semin Immunol. 1999;11(6):375–384. - PubMed
-
- Storkus WJ, Zeh HJ, 3rd, Salter RD, Lotze MT. Identification of T-cell epitopes: rapid isolation of class I-presented peptides from viable cells by mild acid elution. J Immunother Emphasis Tumor Immunol. 1993;14(2):94–103. - PubMed
-
- Torabi-Pour N, Nouri AM, Saffie R, Oliver RT. Comparative study between direct mild acid extraction and immunobead purification technique for isolation of HLA class I-associated peptides. Urol Int. 2002;68(1):38–43. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

