Importance of each residue within secretin for receptor binding and biological activity
- PMID: 21388146
- PMCID: PMC3071462
- DOI: 10.1021/bi200133u
Importance of each residue within secretin for receptor binding and biological activity
Abstract
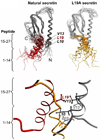
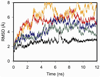
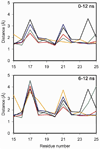
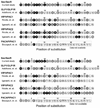
Secretin is a linear 27-residue peptide hormone that stimulates pancreatic and biliary ductular bicarbonate and water secretion by acting at its family B G protein-coupled receptor. While, like other family members, the carboxyl-terminal region of secretin is most important for high affinity binding and its amino-terminal region is most important for receptor selectivity and receptor activation, determinants for these activities are distributed throughout the entire length of this peptide. In this work, we have systematically investigated changing each residue within secretin to alanine and evaluating the impact on receptor binding and biological activity. The residues most critical for receptor binding were His1, Asp3, Gly4, Phe6, Thr7, Ser8, Leu10, Asp15, Leu19, and Leu23. The residues most critical for biological activity included His1, Gly4, Thr7, Ser8, Glu9, Leu10, Leu19, Leu22, and Leu23, with Asp3, Phe6, Ser11, Leu13, Asp15, Leu26, and Val27 also contributing. While the importance of residues in positions analogous to His1, Asp3, Phe6, Thr7, and Leu23 is conserved for several closely related members of this family, Leu19 is uniquely important for secretin. We, therefore, have further studied this residue by molecular modeling and molecular dynamics simulations. Indeed, the molecular dynamics simulations showed that mutation of Leu19 to alanine was destabilizing, with this effect greater than that observed for the analogous position in the other close family members. This could reflect reduced contact with the receptor or an increase in the solvent-accessible surface area of the hydrophobic residues in the carboxyl terminus of secretin as bound to its receptor.
Figures




Similar articles
-
Use of Cysteine Trapping to Map Spatial Approximations between Residues Contributing to the Helix N-capping Motif of Secretin and Distinct Residues within Each of the Extracellular Loops of Its Receptor.J Biol Chem. 2016 Mar 4;291(10):5172-84. doi: 10.1074/jbc.M115.706010. Epub 2016 Jan 6. J Biol Chem. 2016. PMID: 26740626 Free PMC article.
-
Molecular basis of secretin docking to its intact receptor using multiple photolabile probes distributed throughout the pharmacophore.J Biol Chem. 2011 Jul 8;286(27):23888-99. doi: 10.1074/jbc.M111.245969. Epub 2011 May 12. J Biol Chem. 2011. PMID: 21566140 Free PMC article.
-
Lactam constraints provide insights into the receptor-bound conformation of secretin and stabilize a receptor antagonist.Biochemistry. 2011 Sep 27;50(38):8181-92. doi: 10.1021/bi2008036. Epub 2011 Aug 30. Biochemistry. 2011. PMID: 21851058 Free PMC article.
-
Ligand binding and activation of the secretin receptor, a prototypic family B G protein-coupled receptor.Br J Pharmacol. 2012 May;166(1):18-26. doi: 10.1111/j.1476-5381.2011.01463.x. Br J Pharmacol. 2012. PMID: 21542831 Free PMC article. Review.
-
Structural basis of natural ligand binding and activation of the Class II G-protein-coupled secretin receptor.Biochem Soc Trans. 2007 Aug;35(Pt 4):709-12. doi: 10.1042/BST0350709. Biochem Soc Trans. 2007. PMID: 17635130 Review.
Cited by
-
Domain coupling in GPCRs: the engine for induced conformational changes.Trends Pharmacol Sci. 2012 Feb;33(2):79-88. doi: 10.1016/j.tips.2011.09.007. Epub 2011 Oct 29. Trends Pharmacol Sci. 2012. PMID: 22037017 Free PMC article. Review.
-
Molecular basis of peptide activation of the GLP-1 receptor.Mol Metab. 2013 Mar 6;2(2):60-1. doi: 10.1016/j.molmet.2013.02.003. eCollection 2013. Mol Metab. 2013. PMID: 24199149 Free PMC article. No abstract available.
-
Machine-Learning-Guided Peptide Drug Discovery: Development of GLP-1 Receptor Agonists with Improved Drug Properties.J Med Chem. 2024 Jul 25;67(14):11814-11826. doi: 10.1021/acs.jmedchem.4c00417. Epub 2024 Jul 8. J Med Chem. 2024. PMID: 38977267 Free PMC article.
-
Structure and dynamics of the active Gs-coupled human secretin receptor.Nat Commun. 2020 Aug 18;11(1):4137. doi: 10.1038/s41467-020-17791-4. Nat Commun. 2020. PMID: 32811827 Free PMC article.
-
Transmembrane signal transduction by peptide hormones via family B G protein-coupled receptors.Front Pharmacol. 2015 Nov 5;6:264. doi: 10.3389/fphar.2015.00264. eCollection 2015. Front Pharmacol. 2015. PMID: 26594176 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

