Computational design of the sequence and structure of a protein-binding peptide
- PMID: 21388199
- PMCID: PMC3081598
- DOI: 10.1021/ja110296z
Computational design of the sequence and structure of a protein-binding peptide
Abstract
The de novo design of protein-binding peptides is challenging because it requires the identification of both a sequence and a backbone conformation favorable for binding. We used a computational strategy that iterates between structure and sequence optimization to redesign the C-terminal portion of the RGS14 GoLoco motif peptide so that it adopts a new conformation when bound to Gα(i1). An X-ray crystal structure of the redesigned complex closely matches the computational model, with a backbone root-mean-square deviation of 1.1 Å.
Figures


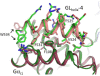
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

