Quantitative miRNA expression analysis using fluidigm microfluidics dynamic arrays
- PMID: 21388556
- PMCID: PMC3062620
- DOI: 10.1186/1471-2164-12-144
Quantitative miRNA expression analysis using fluidigm microfluidics dynamic arrays
Abstract
Background: MicroRNAs (miRNAs) represent a growing class of small non-coding RNAs that are important regulators of gene expression in both plants and animals. Studies have shown that miRNAs play a critical role in human cancer and they can influence the level of cell proliferation and apoptosis by modulating gene expression. Currently, methods for the detection and measurement of miRNA expression include small and moderate-throughput technologies, such as standard quantitative PCR and microarray based analysis. However, these methods have several limitations when used in large clinical studies where a high-throughput and highly quantitative technology needed for the efficient characterization of a large number of miRNA transcripts in clinical samples. Furthermore, archival formalin fixed, paraffin embedded (FFPE) samples are increasingly becoming the primary resource for gene expression studies because fresh frozen (FF) samples are often difficult to obtain and requires special storage conditions. In this study, we evaluated the miRNA expression levels in FFPE and FF samples as well as several lung cancer cell lines employing a high throughput qPCR-based microfluidic technology. The results were compared to standard qPCR and hybridization-based microarray platforms using the same samples.
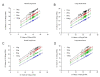
Results: We demonstrated highly correlated Ct values between multiplex and singleplex RT reactions in standard qPCR assays for miRNA expression using total RNA from A549 (R = 0.98; p < 0.0001) and H1299 (R = 0.95; p < 0.0001) lung cancer cell lines. The Ct values generated by the microfluidic technology (Fluidigm 48.48 dynamic array systems) resulted in a left-shift toward lower Ct values compared to those observed by ABI 7900 HT (mean difference, 3.79), suggesting that the microfluidic technology exhibited a greater sensitivity. In addition, we show that as little as 10 ng total RNA can be used to reliably detect all 48 or 96 tested miRNAs using a 96-multiplexing RT reaction in both FFPE and FF samples. Finally, we compared miRNA expression measurements in both FFPE and FF samples by qPCR using the 96.96 dynamic array and Affymetrix microarrays. Fold change comparisons for comparable genes between the two platforms indicated that the overall correlation was R = 0.60. The maximum fold change detected by the Affymetrix microarray was 3.5 compared to 13 by the 96.96 dynamic array.
Conclusion: The qPCR-array based microfluidic dynamic array platform can be used in conjunction with multiplexed RT reactions for miRNA gene expression profiling. We showed that this approach is highly reproducible and the results correlate closely with the existing singleplex qPCR platform at a throughput that is 5 to 20 times higher and a sample and reagent usage that was approximately 50-100 times lower than conventional assays. We established optimal conditions for using the Fluidigm microfluidic technology for rapid, cost effective, and customizable arrays for miRNA expression profiling and validation.
Figures
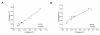



References
-
- Bartel DP, Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nature reviews. 2004;5(5):396–400. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

