Solution structures of Ca2+-CIB1 and Mg2+-CIB1 and their interactions with the platelet integrin alphaIIb cytoplasmic domain
- PMID: 21388953
- PMCID: PMC3089561
- DOI: 10.1074/jbc.M110.179028
Solution structures of Ca2+-CIB1 and Mg2+-CIB1 and their interactions with the platelet integrin alphaIIb cytoplasmic domain
Abstract
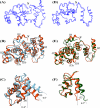
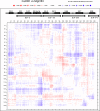
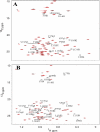
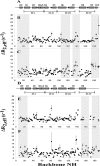
The calcium- and integrin-binding protein 1 (CIB1) is a ubiquitous Ca(2+)-binding protein and a specific binding partner for the platelet integrin αIIb cytoplasmic domain, which confers the key role of CIB1 in hemostasis. CIB1 is also known to be involved in apoptosis, embryogenesis, and the DNA damage response. In this study, the solution structures of both Ca(2+)-CIB1 and Mg(2+)-CIB1 were determined using solution-state NMR spectroscopy. The methyl groups of Ile, Leu, and Val were selectively protonated to compensate for the loss of protons due to deuteration. The solution structure of Ca(2+)-CIB1 possesses smaller opened EF-hands in its C-domain compared with available crystal structures. Ca(2+)-CIB1 and Mg(2+)-CIB1 have similar structures, but the N-lobe of Mg(2+)-CIB1 is slightly more opened than that of Ca(2+)-CIB1. Additional NMR experiments, such as chemical shift perturbation and methyl group solvent accessibility as measured by a nitroxide surface probe, were carried out to further characterize the structures of Ca(2+)-CIB1 and Mg(2+)-CIB1 as well as their interactions with the integrin αIIb cytoplasmic domain. NMR measurements of backbone amide proton slow motion (microsecond to millisecond) dynamics confirmed that the C-terminal helix of Ca(2+)-CIB1 is displaced upon αIIb binding. The EF-hand III of both Ca(2+)-CIB1 and Mg(2+)-CIB1 was identified to be directly involved in the interaction of CIB1 with αIIb. Together, these data illustrate that CIB1 behaves quite differently from related EF-hand regulatory calcium-binding proteins, such as calmodulin or neuronal calcium sensor proteins.
Figures



References
-
- Gifford J. L., Walsh M. P., Vogel H. J. (2007) Biochem. J. 405, 199–221 - PubMed
-
- Yamniuk A. P., Vogel H. J. (2006) Calcium Bind. Proteins 1, 150–155
-
- Leisner T. M., Yuan W., DeNofrio J. C., Liu J., Parise L. V. (2007) Curr. Opin. Hematol. 14, 255–261 - PubMed
-
- Naik U. P., Patel P. M., Parise L. V. (1997) J. Biol. Chem. 272, 4651–4654 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

