Roles of chromatin remodeling factors in the formation and maintenance of heterochromatin structure
- PMID: 21388962
- PMCID: PMC3077663
- DOI: 10.1074/jbc.M110.183269
Roles of chromatin remodeling factors in the formation and maintenance of heterochromatin structure
Abstract
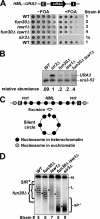
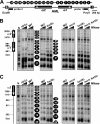
Heterochromatin consists of highly ordered nucleosomes with characteristic histone modifications. There is evidence implicating chromatin remodeling proteins in heterochromatin formation, but their exact roles are not clear. We demonstrate in Saccharomyces cerevisiae that the Fun30p and Isw1p chromatin remodeling factors are similarly required for transcriptional silencing at the HML locus, but they differentially contribute to the structure and stability of HML heterochromatin. In the absence of Fun30p, only a partially silenced structure is established at HML. Such a structure resembles fully silenced heterochromatin in histone modifications but differs markedly from both fully silenced and derepressed chromatin structures regarding nucleosome arrangement. This structure likely represents an intermediate state of heterochromatin that can be converted by Fun30p to the mature state. Moreover, Fun30p removal reduces the rate of de novo establishment of heterochromatin, suggesting that Fun30p assists the silencing machinery in forming heterochromatin. We also find evidence suggesting that Fun30p functions together with, or after, the action of the silencing machinery. On the other hand, Isw1p is dispensable for the formation of heterochromatin structure but is instead critically required for maintaining its stability. Therefore, chromatin remodeling proteins may rearrange nucleosomes during the formation of heterochromatin or serve to stabilize/maintain heterochromatin structure.
Figures







Similar articles
-
Functions of protosilencers in the formation and maintenance of heterochromatin in Saccharomyces cerevisiae.PLoS One. 2012;7(5):e37092. doi: 10.1371/journal.pone.0037092. Epub 2012 May 17. PLoS One. 2012. PMID: 22615905 Free PMC article.
-
A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin.PLoS Biol. 2004 May;2(5):E131. doi: 10.1371/journal.pbio.0020131. Epub 2004 Mar 23. PLoS Biol. 2004. PMID: 15045029 Free PMC article.
-
Differential contributions of histone H3 and H4 residues to heterochromatin structure.Genetics. 2011 Jun;188(2):291-308. doi: 10.1534/genetics.111.127886. Epub 2011 Mar 24. Genetics. 2011. PMID: 21441216 Free PMC article.
-
Keeping chromatin quiet: how nucleosome remodeling restores heterochromatin after replication.Cell Cycle. 2011 Dec 1;10(23):4017-25. doi: 10.4161/cc.10.23.18558. Epub 2011 Dec 1. Cell Cycle. 2011. PMID: 22101266 Free PMC article. Review.
-
Functions of chromatin remodeling factors in heterochromatin formation and maintenance.Sci China Life Sci. 2012 Jan;55(1):89-96. doi: 10.1007/s11427-012-4267-1. Epub 2012 Feb 8. Sci China Life Sci. 2012. PMID: 22314495 Review.
Cited by
-
Functions of Fun30 chromatin remodeler in regulating cellular resistance to genotoxic stress.PLoS One. 2015 Mar 25;10(3):e0121341. doi: 10.1371/journal.pone.0121341. eCollection 2015. PLoS One. 2015. PMID: 25806814 Free PMC article.
-
Nucleosome Positioning Regulates the Establishment, Stability, and Inheritance of Heterochromatin in Saccharomyces cerevisiae.Proc Natl Acad Sci U S A. 2020 Nov 3;117(44):27493-27501. doi: 10.1073/pnas.2004111117. Epub 2020 Oct 19. Proc Natl Acad Sci U S A. 2020. PMID: 33077593 Free PMC article.
-
Crystal structure of the ATPase-C domain of the chromatin remodeller Fun30 from Saccharomyces cerevisiae.Acta Crystallogr F Struct Biol Commun. 2017 Jan 1;73(Pt 1):9-15. doi: 10.1107/S2053230X16019269. Epub 2017 Jan 1. Acta Crystallogr F Struct Biol Commun. 2017. PMID: 28045388 Free PMC article.
-
Determinants of Sir2-Mediated, Silent Chromatin Cohesion.Mol Cell Biol. 2016 Jul 14;36(15):2039-50. doi: 10.1128/MCB.00057-16. Print 2016 Aug 1. Mol Cell Biol. 2016. PMID: 27185881 Free PMC article.
-
Nucleosome Remodeling by Fun30SMARCAD1 in the DNA Damage Response.Front Mol Biosci. 2019 Sep 4;6:78. doi: 10.3389/fmolb.2019.00078. eCollection 2019. Front Mol Biosci. 2019. PMID: 31555662 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

