The survival of motor neuron (SMN) protein interacts with the mRNA-binding protein HuD and regulates localization of poly(A) mRNA in primary motor neuron axons
- PMID: 21389246
- PMCID: PMC3070748
- DOI: 10.1523/JNEUROSCI.3631-10.2011
The survival of motor neuron (SMN) protein interacts with the mRNA-binding protein HuD and regulates localization of poly(A) mRNA in primary motor neuron axons
Abstract
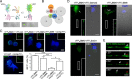
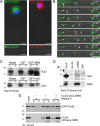
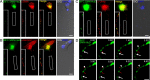
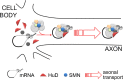
Spinal muscular atrophy (SMA) results from reduced levels of the survival of motor neuron (SMN) protein, which has a well characterized function in spliceosomal small nuclear ribonucleoprotein assembly. Currently, it is not understood how deficiency of a housekeeping protein leads to the selective degeneration of spinal cord motor neurons. Numerous studies have shown that SMN is present in neuronal processes and has many interaction partners, including mRNA-binding proteins, suggesting a potential noncanonical role in axonal mRNA metabolism. In this study, we have established a novel technological approach using bimolecular fluorescence complementation (BiFC) and quantitative image analysis to characterize SMN-protein interactions in primary motor neurons. Consistent with biochemical studies on the SMN complex, BiFC analysis revealed that SMN dimerizes and interacts with Gemin2 in nuclear gems and axonal granules. In addition, using pull down assays, immunofluorescence, cell transfection, and BiFC, we characterized a novel interaction between SMN and the neuronal mRNA-binding protein HuD, which was dependent on the Tudor domain of SMN. A missense mutation in the SMN Tudor domain, which is known to cause SMA, impaired the interaction with HuD, but did not affect SMN axonal localization or self-association. Furthermore, time-lapse microscopy revealed SMN cotransport with HuD in live motor neurons. Importantly, SMN knockdown in primary motor neurons resulted in a specific reduction of both HuD protein and poly(A) mRNA levels in the axonal compartment. These findings reveal a noncanonical role for SMN whereby its interaction with mRNA-binding proteins may facilitate the localization of associated poly(A) mRNAs into axons.
Figures





References
-
- Anthony K, Gallo JM. Aberrant RNA processing events in neurological disorders. Brain Res. 2010;1338:67–77. - PubMed
-
- Aronov S, Aranda G, Behar L, Ginzburg I. Visualization of translated tau protein in the axons of neuronal P19 cells and characterization of tau RNP granules. J Cell Sci. 2002;115:3817–3827. - PubMed
-
- Atlas R, Behar L, Elliott E, Ginzburg I. The insulin-like growth factor mRNA binding-protein IMP-1 and the Ras-regulatory protein G3BP associate with tau mRNA and HuD protein in differentiated P19 neuronal cells. J Neurochem. 2004;89:613–626. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
