The structural basis of agonist-induced activation in constitutively active rhodopsin
- PMID: 21389983
- PMCID: PMC3715716
- DOI: 10.1038/nature09795
The structural basis of agonist-induced activation in constitutively active rhodopsin
Abstract
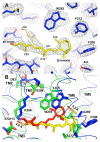
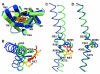
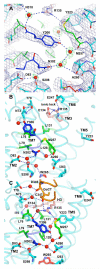
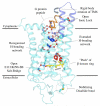
G-protein-coupled receptors (GPCRs) comprise the largest family of membrane proteins in the human genome and mediate cellular responses to an extensive array of hormones, neurotransmitters and sensory stimuli. Although some crystal structures have been determined for GPCRs, most are for modified forms, showing little basal activity, and are bound to inverse agonists or antagonists. Consequently, these structures correspond to receptors in their inactive states. The visual pigment rhodopsin is the only GPCR for which structures exist that are thought to be in the active state. However, these structures are for the apoprotein, or opsin, form that does not contain the agonist all-trans retinal. Here we present a crystal structure at a resolution of 3 Å for the constitutively active rhodopsin mutant Glu 113 Gln in complex with a peptide derived from the carboxy terminus of the α-subunit of the G protein transducin. The protein is in an active conformation that retains retinal in the binding pocket after photoactivation. Comparison with the structure of ground-state rhodopsin suggests how translocation of the retinal β-ionone ring leads to a rotation of transmembrane helix 6, which is the critical conformational change on activation. A key feature of this conformational change is a reorganization of water-mediated hydrogen-bond networks between the retinal-binding pocket and three of the most conserved GPCR sequence motifs. We thus show how an agonist ligand can activate its GPCR.
Figures
References
-
- Park JH, et al. Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature. 2008;454(7201):183. - PubMed
-
- Scheerer P, et al. Crystal structure of opsin in its G-protein-interacting conformation. Nature. 2008;455(7212):497. - PubMed
-
- Zhukovsky EA, Oprian DD. Effect of carboxylic acid side chains on the absorption maximum of visual pigments. Science. 1989;246(4932):928. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

