Inactivating mutations of acetyltransferase genes in B-cell lymphoma
- PMID: 21390126
- PMCID: PMC3271441
- DOI: 10.1038/nature09730
Inactivating mutations of acetyltransferase genes in B-cell lymphoma
Abstract
B-cell non-Hodgkin's lymphoma comprises biologically and clinically distinct diseases the pathogenesis of which is associated with genetic lesions affecting oncogenes and tumour-suppressor genes. We report here that the two most common types--follicular lymphoma and diffuse large B-cell lymphoma--harbour frequent structural alterations inactivating CREBBP and, more rarely, EP300, two highly related histone and non-histone acetyltransferases (HATs) that act as transcriptional co-activators in multiple signalling pathways. Overall, about 39% of diffuse large B-cell lymphoma and 41% of follicular lymphoma cases display genomic deletions and/or somatic mutations that remove or inactivate the HAT coding domain of these two genes. These lesions usually affect one allele, suggesting that reduction in HAT dosage is important for lymphomagenesis. We demonstrate specific defects in acetylation-mediated inactivation of the BCL6 oncoprotein and activation of the p53 tumour suppressor. These results identify CREBBP/EP300 mutations as a major pathogenetic mechanism shared by common forms of B-cell non-Hodgkin's lymphoma, with direct implications for the use of drugs targeting acetylation/deacetylation mechanisms.
Figures
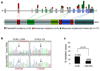

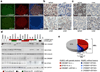


References
-
- Swerdlow SH, et al. Lyon: International Agency for Research on Cancer (IARC); 2008. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues.
-
- Klein U, Dalla-Favera R. Germinal centres: role in B-cell physiology and malignancy. Nat Rev Immunol. 2008;8:22–33. - PubMed
-
- Lenz G, et al. Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science. 2008;319:1676–1679. - PubMed
References not cited in the main text
-
- Bieber T, Elsasser HP. Preparation of a low molecular weight polyethylenimine for efficient cell transfection. Biotechniques. 2001;30:74–77. 80–81. - PubMed
-
- Tang Y, Luo J, Zhang W, Gu W. Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol Cell. 2006;24:827–839. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- U54-AI057158/AI/NIAID NIH HHS/United States
- R01-CA37295/CA/NCI NIH HHS/United States
- DE018183/DE/NIDCR NIH HHS/United States
- R01 CA037295/CA/NCI NIH HHS/United States
- P01-CA092625/CA/NCI NIH HHS/United States
- 1R01LM010140-01/LM/NLM NIH HHS/United States
- R01 LM010140/LM/NLM NIH HHS/United States
- R37 CA037295/CA/NCI NIH HHS/United States
- P01 CA092625/CA/NCI NIH HHS/United States
- R21 DE018183/DE/NIDCR NIH HHS/United States
- P30 CA021765/CA/NCI NIH HHS/United States
- U54 AI057158/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

