Molecular epidemiology and evolution of human respiratory syncytial virus and human metapneumovirus
- PMID: 21390255
- PMCID: PMC3046979
- DOI: 10.1371/journal.pone.0017427
Molecular epidemiology and evolution of human respiratory syncytial virus and human metapneumovirus
Abstract
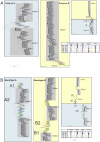
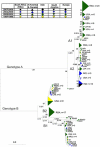
Human respiratory syncytial virus (HRSV) and human metapneumovirus (HMPV) are ubiquitous respiratory pathogens of the Pneumovirinae subfamily of the Paramyxoviridae. Two major surface antigens are expressed by both viruses; the highly conserved fusion (F) protein, and the extremely diverse attachment (G) glycoprotein. Both viruses comprise two genetic groups, A and B. Circulation frequencies of the two genetic groups fluctuate for both viruses, giving rise to frequently observed switching of the predominantly circulating group. Nucleotide sequence data for the F and G gene regions of HRSV and HMPV variants from the UK, The Netherlands, Bangkok and data available from Genbank were used to identify clades of both viruses. Several contemporary circulating clades of HRSV and HMPV were identified by phylogenetic reconstructions. The molecular epidemiology and evolutionary dynamics of clades were modelled in parallel. Times of origin were determined and positively selected sites were identified. Sustained circulation of contemporary clades of both viruses for decades and their global dissemination demonstrated that switching of the predominant genetic group did not arise through the emergence of novel lineages each respiratory season, but through the fluctuating circulation frequencies of pre-existing lineages which undergo proliferative and eclipse phases. An abundance of sites were identified as positively selected within the G protein but not the F protein of both viruses. For HRSV, these were discordant with previously identified residues under selection, suggesting the virus can evade immune responses by generating diversity at multiple sites within linear epitopes. For both viruses, different sites were identified as positively selected between genetic groups.
Conflict of interest statement
Figures

References
-
- Anderson LJ, Hierholzer JC, Tsou C, Hendry RM, Fernie BF, et al. Antigenic characterization of respiratory syncytial virus strains with monoclonal antibodies. J Inf Dis. 1985;151(4):626–633. - PubMed
-
- Mufson MA, Orvell C, Rafnar B, Norrby E. Two distinct subtypes of human respiratory syncytial virus. J Gen Virol. 1985;66(10):2111–2124. - PubMed
-
- McGill A, Greensill J, Marsh R, Craft AW, Toms GL. Detection of human respiratory syncytial virus genotype specific antibody responses in infants. J Med Virol. 2004;74(3):492–498. - PubMed
-
- Venter M, Madhi SA, Tiemessen CT, Schoub BD. Genetic diversity and molecular epidemiology of respiratory syncytial virus over four consecutive seasons in South Africa: identification of new subgroup A and B genotypes. J Gen Virol. 2001;82(9):2117–2124. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

