Amplification of emerging viruses in a bat colony
- PMID: 21392436
- PMCID: PMC3165994
- DOI: 10.3201/eid1703.100526
Amplification of emerging viruses in a bat colony
Abstract
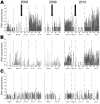
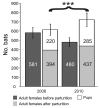
Bats host noteworthy viral pathogens, including coronaviruses, astroviruses, and adenoviruses. Knowledge on the ecology of reservoir-borne viruses is critical for preventive approaches against zoonotic epidemics. We studied a maternity colony of Myotis myotis bats in the attic of a private house in a suburban neighborhood in Rhineland-Palatinate, Germany, during 2008, 2009, and 2010. One coronavirus, 6 astroviruses, and 1 novel adenovirus were identified and monitored quantitatively. Strong and specific amplification of RNA viruses, but not of DNA viruses, occurred during colony formation and after parturition. The breeding success of the colony was significantly better in 2010 than in 2008, in spite of stronger amplification of coronaviruses and astroviruses in 2010, suggesting that these viruses had little pathogenic influence on bats. However, the general correlation of virus and bat population dynamics suggests that bats control infections similar to other mammals and that they may well experience epidemics of viruses under certain circumstances.
Figures


References
-
- Simmons NB. Order Chiroptera. In: Wilson DE, Reeder DM, editors. Mammal species of the world: a taxonomic and geographic reference. Baltimore: Johns Hopkins University Press; 2005. p. 312–529.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical
