Dynamics of ESCRT protein recruitment during retroviral assembly
- PMID: 21394083
- PMCID: PMC3245320
- DOI: 10.1038/ncb2207
Dynamics of ESCRT protein recruitment during retroviral assembly
Abstract
The ESCRT (endosomal sorting complex required for transport) complexes and associated proteins mediate membrane scission reactions, such as multivesicular body formation, the terminal stages of cytokinesis and retroviral particle release. These proteins are believed to be sequentially recruited to the site of membrane scission, and then complexes are disassembled by the ATPase Vps4A. However, these events have never been observed in living cells, and their dynamics are unknown. By quantifying the recruitment of several ESCRT and associated proteins during the assembly of two retroviruses, we show that Alix progressively accumulated at viral assembly sites, coincident with the accumulation of the main viral structural protein, Gag, and was not recycled after assembly. In contrast, ESCRT-III and Vps4A were transiently recruited only when the accumulation of Gag was complete. These data indicate that the rapid and transient recruitment of proteins that act late in the ESCRT pathway and carry out membrane fission is triggered by prior and progressive accumulation of proteins that bridge viral proteins and the late-acting ESCRT proteins.
© 2011 Macmillan Publishers Limited. All rights reserved
Conflict of interest statement
The authors declare no competing financial interests.
Figures
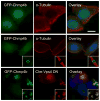
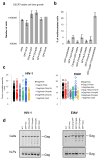
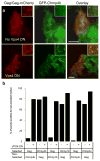


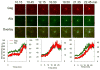

Comment in
-
Virology: ESCRTing retroviruses to the exit.Nat Rev Microbiol. 2011 May;9(5):314. doi: 10.1038/nrmicro2571. Nat Rev Microbiol. 2011. PMID: 21494275 No abstract available.
References
-
- Bieniasz PD. Late budding domains and host proteins in enveloped virus release. Virology. 2006;344:55–63. - PubMed
-
- Morita E, Sundquist WI. Retrovirus budding. Annu Rev Cell Dev Biol. 2004;20:395–425. - PubMed
-
- Williams RL, Urbe S. The emerging shape of the ESCRT machinery. Nat Rev Mol Cell Biol. 2007;8:355–368. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

