Nature of allosteric inhibition in glutamate racemase: discovery and characterization of a cryptic inhibitory pocket using atomistic MD simulations and pKa calculations
- PMID: 21395329
- PMCID: PMC3072873
- DOI: 10.1021/jp201037t
Nature of allosteric inhibition in glutamate racemase: discovery and characterization of a cryptic inhibitory pocket using atomistic MD simulations and pKa calculations
Abstract
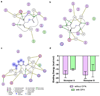
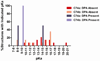
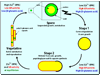
Enzyme inhibition via allostery, in which the ligand binds remotely from the active site, is a poorly understood phenomenon and represents a significant challenge to structure-based drug design. Dipicolinic acid (DPA), a major component of Bacillus spores, is shown to inhibit glutamate racemase from Bacillus anthracis , a monosubstrate/monoproduct enzyme, in a novel allosteric fashion. Glutamate racemase has long been considered an important drug target for its integral role in bacterial cell wall synthesis. The DPA binding mode was predicted via multiple docking studies and validated via site-directed mutagenesis at the binding locus, while the mechanism of inhibition was elucidated with a combination of Blue Native polyacrylamide gel electrophoresis, molecular dynamics simulations, and free energy and pK(a) calculations. Inhibition by DPA not only reveals a novel cryptic binding site but also represents a form of allosteric regulation that exploits the interplay between enzyme conformational changes, fluctuations in the pK(a) values of buried residues and catalysis. The potential for future drug development is discussed.
Figures

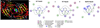


Similar articles
-
Functional comparison of the two Bacillus anthracis glutamate racemases.J Bacteriol. 2007 Jul;189(14):5265-75. doi: 10.1128/JB.00352-07. Epub 2007 May 11. J Bacteriol. 2007. PMID: 17496086 Free PMC article.
-
Exploitation of structural and regulatory diversity in glutamate racemases.Nature. 2007 Jun 14;447(7146):817-22. doi: 10.1038/nature05689. Nature. 2007. PMID: 17568739
-
Glutamate racemase dimerization inhibits dynamic conformational flexibility and reduces catalytic rates.Biochemistry. 2009 Jul 28;48(29):7045-55. doi: 10.1021/bi9005072. Biochemistry. 2009. PMID: 19552402 Free PMC article.
-
Glutamate racemase as a target for drug discovery.Microb Biotechnol. 2008 Sep;1(5):345-60. doi: 10.1111/j.1751-7915.2008.00031.x. Epub 2008 May 11. Microb Biotechnol. 2008. PMID: 21261855 Free PMC article. Review.
-
Biochemistry and molecular genetics of poly-gamma-glutamate synthesis.Appl Microbiol Biotechnol. 2002 Jun;59(1):9-14. doi: 10.1007/s00253-002-0984-x. Epub 2002 Apr 16. Appl Microbiol Biotechnol. 2002. PMID: 12073126 Review.
Cited by
-
Integrating Experimental and In Silico HTS in the Discovery of Inhibitors of Protein-Nucleic Acid Interactions.Methods Enzymol. 2018;601:243-273. doi: 10.1016/bs.mie.2017.11.036. Epub 2018 Feb 28. Methods Enzymol. 2018. PMID: 29523234 Free PMC article.
-
Systematic exploration of multiple drug binding sites.J Cheminform. 2017 Dec 28;9(1):65. doi: 10.1186/s13321-017-0255-6. J Cheminform. 2017. PMID: 29282592 Free PMC article.
-
Racemases and epimerases operating through a 1,1-proton transfer mechanism: reactivity, mechanism and inhibition.Chem Soc Rev. 2021 May 24;50(10):5952-5984. doi: 10.1039/d0cs00540a. Chem Soc Rev. 2021. PMID: 34027955 Free PMC article. Review.
-
Flooding enzymes: quantifying the contributions of interstitial water and cavity shape to ligand binding using extended linear response free energy calculations.J Chem Inf Model. 2013 Sep 23;53(9):2349-59. doi: 10.1021/ci400244x. Epub 2013 Sep 6. J Chem Inf Model. 2013. PMID: 24111836 Free PMC article.
-
MptpA Kinetics Enhanced by Allosteric Control of an Active Conformation.J Mol Biol. 2022 Sep 15;434(17):167540. doi: 10.1016/j.jmb.2022.167540. Epub 2022 Mar 23. J Mol Biol. 2022. PMID: 35339563 Free PMC article.
References
-
- Rogers HJ, Perkins HR, Ward JB. Microbial cell walls and membranes. London, United Kingdom: Chapman and Hall; 1980.
-
- de Dios A, Prieto L, Martin JA, Rubio A, Ezquerra J, Tebbe M, Lopez de Uralde B, Martin J, Sanchez A, LeTourneau DL, McGee JE, Boylan C, Parr TR, Jr, Smith MC. J Med Chem. 2002;45:4559. - PubMed
-
- Glavas S, Tanner ME. Bioorganic and Medicinal Chemistry Letters. 1997;7:2265.
-
- Lundqvist T, Fisher SL, Kern G, Folmer RH, Xue Y, Newton DT, Keating TA, Alm RA, de Jonge BL. Nature. 2007;447:817. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

