Characterization of an in vitro differentiation assay for pancreatic-like cell development from murine embryonic stem cells: detailed gene expression analysis
- PMID: 21395400
- PMCID: PMC3148116
- DOI: 10.1089/adt.2010.0314
Characterization of an in vitro differentiation assay for pancreatic-like cell development from murine embryonic stem cells: detailed gene expression analysis
Abstract
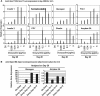
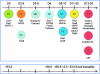
Embryonic stem (ES) cell technology may serve as a platform for the discovery of drugs to treat diseases such as diabetes. However, because of difficulties in establishing reliable ES cell differentiation methods and in creating cost-effective plating conditions for the high-throughput format, screening for molecules that regulate pancreatic beta cells and their immediate progenitors has been limited. A relatively simple and inexpensive differentiation protocol that allows efficient generation of insulin-expressing cells from murine ES cells was previously established in our laboratories. In this report, this system is characterized in greater detail to map developmental cell stages for future screening experiments. Our results show that sequential activation of multiple gene markers for undifferentiated ES cells, epiblast, definitive endoderm, foregut, and pancreatic lineages was found to follow the sequence of events that mimics pancreatic ontogeny. Cells that expressed enhanced green fluorescent protein, driven by pancreatic and duodenal homeobox 1 or insulin 1 promoter, correctly expressed known beta cell lineage markers. Overexpression of Sox17, an endoderm fate-determining transcription factor, at a very early stage of differentiation (days 2-3) enhanced pancreatic gene expression. Overexpression of neurogenin3, an endocrine progenitor cell marker, induced glucagon expression at stages when pancreatic and duodenal homeobox 1 message was present (days 10-16). Forced expression (between days 16 and 25) of MafA, a pancreatic maturation factor, resulted in enhanced expression of insulin genes, glucose transporter 2 and glucokinase, and glucose-responsive insulin secretion. Day 20 cells implanted in vivo resulted in pancreatic-like cells. Together, our differentiation assay recapitulates the proceedings and behaviors of pancreatic development and will be valuable for future screening of beta cell effectors.
Figures









Similar articles
-
Ectopic Ptf1a expression in murine ESCs potentiates endocrine differentiation and models pancreas development in vitro.Stem Cells. 2014 May;32(5):1195-207. doi: 10.1002/stem.1616. Stem Cells. 2014. PMID: 24375815 Free PMC article.
-
Matched miRNA and mRNA signatures from an hESC-based in vitro model of pancreatic differentiation reveal novel regulatory interactions.J Cell Sci. 2013 Sep 1;126(Pt 17):3848-61. doi: 10.1242/jcs.123570. Epub 2013 Jun 26. J Cell Sci. 2013. PMID: 23813959 Free PMC article.
-
Switching of mesodermal and endodermal properties in hTERT-modified and expanded fetal human pancreatic progenitor cells.Stem Cell Res Ther. 2010 Mar 15;1(1):6. doi: 10.1186/scrt6. Stem Cell Res Ther. 2010. PMID: 20504287 Free PMC article.
-
Genes controlling pancreas ontogeny.Int J Dev Biol. 2008;52(7):823-35. doi: 10.1387/ijdb.072444cb. Int J Dev Biol. 2008. PMID: 18956314 Review.
-
In vitro models of pancreatic differentiation using embryonic stem or induced pluripotent stem cells.Congenit Anom (Kyoto). 2011 Mar;51(1):21-5. doi: 10.1111/j.1741-4520.2010.00307.x. Congenit Anom (Kyoto). 2011. PMID: 21129040 Review.
Cited by
-
In vitro multilineage differentiation and self-renewal of single pancreatic colony-forming cells from adult C57BL/6 mice.Stem Cells Dev. 2014 Apr 15;23(8):899-909. doi: 10.1089/scd.2013.0466. Epub 2014 Jan 4. Stem Cells Dev. 2014. PMID: 24261600 Free PMC article.
-
A quantitative assay for insulin-expressing colony-forming progenitors.J Vis Exp. 2011 Nov 28;(57):e3148. doi: 10.3791/3148. J Vis Exp. 2011. PMID: 22143165 Free PMC article.
-
Protein Methyltransferase Inhibition Decreases Endocrine Specification Through the Upregulation of Aldh1b1 Expression.Stem Cells. 2019 May;37(5):640-651. doi: 10.1002/stem.2979. Epub 2019 Feb 13. Stem Cells. 2019. PMID: 30681750 Free PMC article.
-
β-Cell Replacement Strategies: The Increasing Need for a "β-Cell Dogma".Front Genet. 2017 Jun 6;8:75. doi: 10.3389/fgene.2017.00075. eCollection 2017. Front Genet. 2017. PMID: 28634486 Free PMC article. Review.
-
Glucocorticoid Signaling Enhances Expression of Glucose-Sensing Molecules in Immature Pancreatic Beta-Like Cells Derived from Murine Embryonic Stem Cells In Vitro.Stem Cells Dev. 2018 Jul 1;27(13):898-909. doi: 10.1089/scd.2017.0160. Epub 2018 Jun 6. Stem Cells Dev. 2018. PMID: 29717618 Free PMC article.
References
-
- Barbaric I. Gokhale PJ. Andrews PW. High-content screening of small compounds on human embryonic stem cells. Biochem Soc Trans. 2010;38:1046–1050. - PubMed
-
- Zhu S. Wurdak H. Wang J. Lyssiotis CA. Peters EC. Cho CY. Wu X. Schultz PG. A small molecule primes embryonic stem cells for differentiation. Cell Stem Cell. 2009;4:416–426. - PubMed
-
- Barbaric I. Gokhale PJ. Jones M. Glen A. Baker D. Andrews PW. Novel regulators of stem cell fates identified by a multivariate phenotype screen of small compounds on human embryonic stem cell colonies. Stem Cell Res. 2010;5:104–119. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
