A recurrent deletion of DPY19L2 causes infertility in man by blocking sperm head elongation and acrosome formation
- PMID: 21397064
- PMCID: PMC3059422
- DOI: 10.1016/j.ajhg.2011.02.007
A recurrent deletion of DPY19L2 causes infertility in man by blocking sperm head elongation and acrosome formation
Abstract
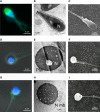
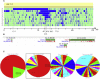
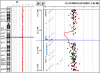
An increasing number of couples require medical assistance to achieve a pregnancy, and more than 2% of the births in Western countries now result from assisted reproductive technologies. To identify genetic variants responsible for male infertility, we performed a whole-genome SNP scan on patients presenting with total globozoospermia, a primary infertility phenotype characterized by the presence of 100% round acrosomeless spermatozoa in the ejaculate. This strategy allowed us to identify in most patients (15/20) a 200 kb homozygous deletion encompassing only DPY19L2, which is highly expressed in the testis. Although there was no known function for DPY19L2 in humans, previous work indicated that its ortholog in C. elegans is involved in cell polarity. In man, the DPY19L2 region has been described as a copy-number variant (CNV) found to be duplicated and heterozygously deleted in healthy individuals. We show here that the breakpoints of the deletions are located on a highly homologous 28 kb low copy repeat (LCR) sequence present on each side of DPY19L2, indicating that the identified deletions were probably produced by nonallelic homologous recombination (NAHR) between these two regions. We demonstrate that patients with globozoospermia have a homozygous deletion of DPY19L2, thus indicating that DPY19L2 is necessary in men for sperm head elongation and acrosome formation. A molecular diagnosis can now be proposed to affected men; the presence of the deletion confirms the diagnosis of globozoospermia and assigns a poor prognosis for the success of in vitro fertilization.
Copyright © 2011 The American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures


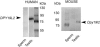
References
-
- Auger J., Kunstmann J.M., Czyglik F., Jouannet P. Decline in semen quality among fertile men in Paris during the past 20 years. N. Engl. J. Med. 1995;332:281–285. - PubMed
-
- Dam A.H., Feenstra I., Westphal J.R., Ramos L., van Golde R.J., Kremer J.A. Globozoospermia revisited. Hum. Reprod. Update. 2007;13:63–75. - PubMed
-
- Kullander S., Rausing A. On round-headed human spermatozoa. Int. J. Fertil. 1975;20:33–40. - PubMed
-
- Flörke-Gerloff S., Töpfer-Petersen E., Müller-Esterl W., Mansouri A., Schatz R., Schirren C., Schill W., Engel W. Biochemical and genetic investigation of round-headed spermatozoa in infertile men including two brothers and their father. Andrologia. 1984;16:187–202. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

