Autosomal-recessive posterior microphthalmos is caused by mutations in PRSS56, a gene encoding a trypsin-like serine protease
- PMID: 21397065
- PMCID: PMC3059417
- DOI: 10.1016/j.ajhg.2011.02.006
Autosomal-recessive posterior microphthalmos is caused by mutations in PRSS56, a gene encoding a trypsin-like serine protease
Abstract
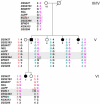
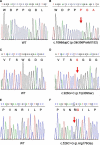
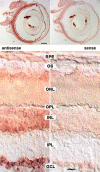
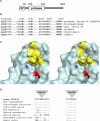
Posterior microphthalmos (MCOP) is a rare isolated developmental anomaly of the eye characterized by extreme hyperopia due to short axial length. The population of the Faroe Islands shows a high prevalence of an autosomal-recessive form (arMCOP) of the disease. Based on published linkage data, we refined the position of the disease locus (MCOP6) in an interval of 250 kb in chromosome 2q37.1 in two large Faroese families. We detected three different mutations in PRSS56. Patients of the Faroese families were either homozygous for c.926G>C (p.Trp309Ser) or compound heterozygous for c.926G>C and c.526C>G (p.Arg176Gly), whereas a homozygous 1 bp duplication (c.1066dupC) was identified in five patients with arMCOP from a consanguineous Tunisian family. In one patient with MCOP from the Faroe Islands and in another one from Turkey, no PRSS56 mutation was detected, suggesting nonallelic heterogeneity of the trait. Using RT-PCR, PRSS56 transcripts were detected in samples derived from the human adult retina, cornea, sclera, and optic nerve. The expression of the mouse ortholog could be first detected in the eye at E17 and was maintained into adulthood. The predicted PRSS56 protein is a 603 amino acid long secreted trypsin-like serine peptidase. The c.1066dupC is likely to result in a functional null allele, whereas the two point mutations predict the replacement of evolutionary conserved and functionally important residues. Molecular modeling of the p.Trp309Ser mutant suggests that both the affinity and reactivity of the enzyme toward in vivo protein substrates are likely to be substantially reduced.
Copyright © 2011 The American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures

References
-
- Fuchs J., Holm K., Vilhelmsen K., Rosenberg T., Scherfig E., Fledelius H.C. Hereditary high hypermetropia in the Faroe Islands. Ophthalmic Genet. 2005;26:9–15. - PubMed
-
- Hmani-Aifa M., Ben Salem S., Benzina Z., Bouassida W., Messaoud R., Turki K., Khairallah M., Rebaï A., Fakhfekh F., Söderkvist P., Ayadi H. A genome-wide linkage scan in Tunisian families identifies a novel locus for non-syndromic posterior microphthalmia to chromosome 2q37.1. Hum. Genet. 2009;126:575–587. - PubMed
-
- Fledelius H.C., Fuchs H.J., Rosenberg T. Oculometric characteristics of extreme hypermetropia in two faroese families. Optom. Vis. Sci. 2004;81:762–768. - PubMed
-
- Walsh M.K., Goldberg M.F. Abnormal foveal avascular zone in nanophthalmos. Am. J. Ophthalmol. 2007;143:1067–1068. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

