Cooperativity within and among Pten, p53, and Rb pathways induces high-grade astrocytoma in adult brain
- PMID: 21397855
- PMCID: PMC3060664
- DOI: 10.1016/j.ccr.2011.01.039
Cooperativity within and among Pten, p53, and Rb pathways induces high-grade astrocytoma in adult brain
Abstract
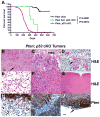
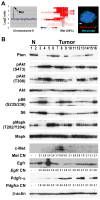
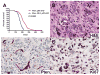
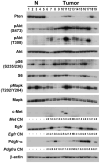
Mutations in the PTEN, TP53, and RB1 pathways are obligate events in the pathogenesis of human glioblastomas. We induced various combinations of deletions in these tumor suppressors in astrocytes and neural precursors in mature mice, resulting in astrocytomas ranging from grade III to grade IV (glioblastoma). There was selection for mutation of multiple genes within a pathway, shown by somatic amplifications of genes in the PI3K or Rb pathway in tumors in which Pten or Rb deletion was an initiating event. Despite multiple mutations within PI3K and Rb pathways, elevated Mapk activation was not consistent. Gene expression profiling revealed striking similarities to subclasses of human diffuse astrocytoma. Astrocytomas were found within and outside of proliferative niches in the adult brain.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures



Comment in
-
Glioma models: new GEMMs add "class" with genomic and expression correlations.Cancer Cell. 2011 Mar 8;19(3):295-7. doi: 10.1016/j.ccr.2011.02.018. Cancer Cell. 2011. PMID: 21397851
References
-
- Backman SA, Stambolic V, Suzuki A, Haight J, Elia A, Pretorius J, Tsao MS, Shannon P, Bolon B, Ivy GO, Mak TW. Deletion of Pten in mouse brain causes seizures, ataxia and defects in soma size resembling Lhermitte-Duclos disease. Nat Genet. 2001;29:396–403. - PubMed
-
- Brustle O, McKay RD. The neuroepithelial stem cell concept: implications for neuro-oncology. J Neurooncol. 1995;24:57–59. - PubMed
-
- Cengiz B, Gunduz M, Nagatsuka H, Beder L, Gunduz E, Tamamura R, Mahmut N, Fukushima K, Ali MA, Naomoto Y, et al. Fine deletion mapping of chromosome 2q21–37 shows three preferentially deleted regions in oral cancer. Oral Oncol. 2007;43:241–247. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

