Non-identity-mediated CRISPR-bacteriophage interaction mediated via the Csy and Cas3 proteins
- PMID: 21398535
- PMCID: PMC3133329
- DOI: 10.1128/JB.01411-10
Non-identity-mediated CRISPR-bacteriophage interaction mediated via the Csy and Cas3 proteins
Abstract
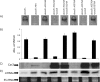
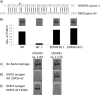
Studies of the Escherichia, Neisseria, Thermotoga, and Mycobacteria clustered regularly interspaced short palindromic repeat (CRISPR) subtypes have resulted in a model whereby CRISPRs function as a defense system against bacteriophage infection and conjugative plasmid transfer. In contrast, we previously showed that the Yersinia-subtype CRISPR region of Pseudomonas aeruginosa strain UCBPP-PA14 plays no detectable role in viral immunity but instead is required for bacteriophage DMS3-dependent inhibition of biofilm formation by P. aeruginosa. The goal of this study is to define the components of the Yersinia-subtype CRISPR region required to mediate this bacteriophage-host interaction. We show that the Yersinia-subtype-specific CRISPR-associated (Cas) proteins Csy4 and Csy2 are essential for small CRISPR RNA (crRNA) production in vivo, while the Csy1 and Csy3 proteins are not absolutely required for production of these small RNAs. Further, we present evidence that the core Cas protein Cas3 functions downstream of small crRNA production and that this protein requires functional HD (predicted phosphohydrolase) and DEXD/H (predicted helicase) domains to suppress biofilm formation in DMS3 lysogens. We also determined that only spacer 1, which is not identical to any region of the DMS3 genome, mediates the CRISPR-dependent loss of biofilm formation. Our evidence suggests that gene 42 of phage DMS3 (DMS3-42) is targeted by CRISPR2 spacer 1 and that this targeting tolerates multiple point mutations between the spacer and DMS3-42 target sequence. This work demonstrates how the interaction between P. aeruginosa strain UCBPP-PA14 and bacteriophage DMS3 can be used to further our understanding of the diverse roles of CRISPR system function in bacteria.
Figures
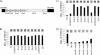





Comment in
-
DMS3-42: the secret to CRISPR-dependent biofilm inhibition in Pseudomonas aeruginosa.J Bacteriol. 2011 Jul;193(14):3431-2. doi: 10.1128/JB.05066-11. Epub 2011 May 6. J Bacteriol. 2011. PMID: 21551309 Free PMC article. No abstract available.
References
-
- Barrangou R., et al. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712 - PubMed
-
- Cady K. C., et al. 2011. Prevalence, conservation and functional analysis of Yersinia and Escherichia CRISPR regions in clinical Pseudomonas aeruginosa isolates. Microbiology 157:430–437 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

