Novel asymmetrically localizing components of human centrosomes identified by complementary proteomics methods
- PMID: 21399614
- PMCID: PMC3102290
- DOI: 10.1038/emboj.2011.63
Novel asymmetrically localizing components of human centrosomes identified by complementary proteomics methods
Abstract
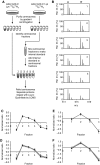
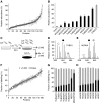
Centrosomes in animal cells are dynamic organelles with a proteinaceous matrix of pericentriolar material assembled around a pair of centrioles. They organize the microtubule cytoskeleton and the mitotic spindle apparatus. Mature centrioles are essential for biogenesis of primary cilia that mediate key signalling events. Despite recent advances, the molecular basis for the plethora of processes coordinated by centrosomes is not fully understood. We have combined protein identification and localization, using PCP-SILAC mass spectrometry, BAC transgeneOmics, and antibodies to define the constituents of human centrosomes. From a background of non-specific proteins, we distinguished 126 known and 40 candidate centrosomal proteins, of which 22 were confirmed as novel components. An antibody screen covering 4000 genes revealed an additional 113 candidates. We illustrate the power of our methods by identifying a novel set of five proteins preferentially associated with mother or daughter centrioles, comprising genes implicated in cell polarity. Pulsed labelling demonstrates a remarkable variation in the stability of centrosomal protein complexes. These spatiotemporal proteomics data provide leads to the further functional characterization of centrosomal proteins.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures





References
-
- Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M (2003) Proteomic characterization of the human centrosome by protein correlation profiling. Nature 426: 570–574 - PubMed
-
- Augustin A, Spenlehauer C, Dumond H, Menissier-De Murcia J, Piel M, Schmit AC, Apiou F, Vonesch JL, Kock M, Bornens M, De Murcia G (2003) PARP-3 localizes preferentially to the daughter centriole and interferes with the G1/S cell cycle progression. J Cell Sci 116: 1551–1562 - PubMed
-
- Barbe L, Lundberg E, Oksvold P, Stenius A, Lewin E, Bjorling E, Asplund A, Ponten F, Brismar H, Uhlen M, Andersson-Svahn H (2008) Toward a confocal subcellular atlas of the human proteome. Mol Cell Proteomics 7: 499–508 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

