Genome-wide association study identifies susceptibility loci for IgA nephropathy
- PMID: 21399633
- PMCID: PMC3412515
- DOI: 10.1038/ng.787
Genome-wide association study identifies susceptibility loci for IgA nephropathy
Abstract
We carried out a genome-wide association study of IgA nephropathy, a major cause of kidney failure worldwide. We studied 1,194 cases and 902 controls of Chinese Han ancestry, with targeted follow up in Chinese and European cohorts comprising 1,950 cases and 1,920 controls. We identified three independent loci in the major histocompatibility complex, as well as a common deletion of CFHR1 and CFHR3 at chromosome 1q32 and a locus at chromosome 22q12 that each surpassed genome-wide significance (P values for association between 1.59 × 10⁻²⁶ and 4.84 × 10⁻⁹ and minor allele odds ratios of 0.63-0.80). These five loci explain 4-7% of the disease variance and up to a tenfold variation in interindividual risk. Many of the alleles that protect against IgA nephropathy impart increased risk for other autoimmune or infectious diseases, and IgA nephropathy risk allele frequencies closely parallel the variation in disease prevalence among Asian, European and African populations, suggesting complex selective pressures.
Figures
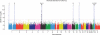


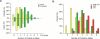
References
-
- Coresh J, et al. Prevalence of chronic kidney disease in the United States. JAMA. 2007;298:2038–47. - PubMed
-
- Tsukamoto Y, et al. Report of the Asian Forum of Chronic Kidney Disease Initiative (AFCKDI) 2007. “Current status and perspective of CKD in Asia”: diversity and specificity among Asian countries. Clin Exp Nephrol. 2009;13:249–56. - PubMed
-
- Gesualdo L, Di Palma AM, Morrone LF, Strippoli GF, Schena FP. The Italian experience of the national registry of renal biopsies. Kidney Int. 2004;66:890–4. - PubMed
-
- D'Amico G. The commonest glomerulonephritis in the world: IgA nephropathy. Q J Med. 1987;64:709–27. - PubMed
-
- Nair R, Walker PD. Is IgA nephropathy the commonest primary glomerulopathy among young adults in the USA? Kidney Int. 2006;69:1455–8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

