Successful targeting and disruption of an integrated reporter lentivirus using the engineered homing endonuclease Y2 I-AniI
- PMID: 21399673
- PMCID: PMC3036713
- DOI: 10.1371/journal.pone.0016825
Successful targeting and disruption of an integrated reporter lentivirus using the engineered homing endonuclease Y2 I-AniI
Abstract
Current antiviral therapy does not cure HIV-infected individuals because the virus establishes lifelong latent infection within long-lived memory T cells as integrated HIV proviral DNA. Here, we report a new therapeutic approach that aims to cure cells of latent HIV infection by rendering latent virus incapable of replication and pathogenesis via targeted cellular mutagenesis of essential viral genes. This is achieved by using a homing endonuclease to introduce DNA double-stranded breaks (dsb) within the integrated proviral DNA, which is followed by triggering of the cellular DNA damage response and error-prone repair. To evaluate this concept, we developed an in vitro culture model of viral latency, consisting of an integrated lentiviral vector with an easily evaluated reporter system to detect targeted mutagenesis events. Using this system, we demonstrate that homing endonucleases can efficiently and selectively target an integrated reporter lentivirus within the cellular genome, leading to mutation in the proviral DNA and loss of reporter gene expression. This new technology offers the possibility of selectively disabling integrated HIV provirus within latently infected cells.
Conflict of interest statement
Figures
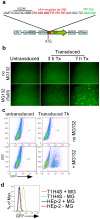

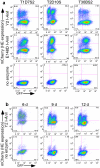

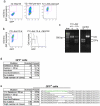




References
-
- Siliciano JD, Lai J, Callender M, Pitt E, Zhang H, et al. Stability of the latent reservoir for HIV-1 in patients receiving valproic acid. J Infect Dis. 2007;195:833–836. - PubMed
-
- Paques F, Duchateau P. Meganucleases and DNA double-strand break-induced recombination: perspectives for gene therapy. Curr Gene Ther. 2007;7:49–66. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

