Resveratrol inhibits the formation of multiple-layered β-sheet oligomers of the human islet amyloid polypeptide segment 22-27
- PMID: 21402038
- PMCID: PMC3059578
- DOI: 10.1016/j.bpj.2011.02.010
Resveratrol inhibits the formation of multiple-layered β-sheet oligomers of the human islet amyloid polypeptide segment 22-27
Erratum in
- Biophys J. 2011 Apr 20;100(8):2076
Abstract
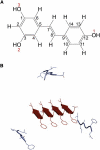
The abnormal self-assembly of a number of proteins or peptides is a hallmark of >20 amyloidogenic diseases. Recent studies suggest that the pathology of amyloidogenesis can be attributed primarily to cytotoxic, soluble, intermediate oligomeric species rather than to mature amyloid fibrils. Despite the lack of available structural information regarding these transient species, many therapeutic efforts have focused on inhibiting the formation of these aggregates. One of the most successful approaches has been to use small molecules, many of which have been found to inhibit toxic species with high efficacy. A significant issue that remains to be resolved is the mechanism underlying the inhibitory effects of these molecules. In this article, we present extensive replica-exchange molecular dynamics simulations to study the early aggregation of the human islet amyloid polypeptide segment 22-27 in the presence and absence of the small-molecule inhibitor resveratrol. The simulations indicate that aggregation of these peptides was hindered by resveratrol via a mechanism of blocking the lateral growth of a single-layered β-sheet oligomer (rather than preventing growth by elongation along the fibril axis). Intersheet side-chain stacking, especially stacking of the aromatic rings, was blocked by the presence of resveratrol molecules, and the overall aggregation level was reduced.
Copyright © 2011 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures






Similar articles
-
Nucleation of β-rich oligomers and β-barrels in the early aggregation of human islet amyloid polypeptide.Biochim Biophys Acta Mol Basis Dis. 2019 Feb 1;1865(2):434-444. doi: 10.1016/j.bbadis.2018.11.021. Epub 2018 Nov 28. Biochim Biophys Acta Mol Basis Dis. 2019. PMID: 30502402 Free PMC article.
-
Peptides Composed of Alternating L- and D-Amino Acids Inhibit Amyloidogenesis in Three Distinct Amyloid Systems Independent of Sequence.J Mol Biol. 2016 Jun 5;428(11):2317-2328. doi: 10.1016/j.jmb.2016.03.013. Epub 2016 Mar 21. J Mol Biol. 2016. PMID: 27012425 Free PMC article.
-
Norepinephrine Inhibits Alzheimer's Amyloid-β Peptide Aggregation and Destabilizes Amyloid-β Protofibrils: A Molecular Dynamics Simulation Study.ACS Chem Neurosci. 2019 Mar 20;10(3):1585-1594. doi: 10.1021/acschemneuro.8b00537. Epub 2019 Jan 15. ACS Chem Neurosci. 2019. PMID: 30605312
-
Aggregation of Aβ(17-36) in the Presence of Naturally Occurring Phenolic Inhibitors Using Coarse-Grained Simulations.J Mol Biol. 2017 Dec 8;429(24):3893-3908. doi: 10.1016/j.jmb.2017.10.006. Epub 2017 Oct 13. J Mol Biol. 2017. PMID: 29031698
-
Understanding amyloid fibril nucleation and aβ oligomer/drug interactions from computer simulations.Acc Chem Res. 2014 Feb 18;47(2):603-11. doi: 10.1021/ar4002075. Epub 2013 Dec 24. Acc Chem Res. 2014. PMID: 24368046 Review.
Cited by
-
Exploring the influence of EGCG on the β-sheet-rich oligomers of human islet amyloid polypeptide (hIAPP1-37) and identifying its possible binding sites from molecular dynamics simulation.PLoS One. 2014 Apr 16;9(4):e94796. doi: 10.1371/journal.pone.0094796. eCollection 2014. PLoS One. 2014. PMID: 24739876 Free PMC article.
-
Amylin uncovered: a review on the polypeptide responsible for type II diabetes.Biomed Res Int. 2013;2013:826706. doi: 10.1155/2013/826706. Epub 2013 Mar 31. Biomed Res Int. 2013. PMID: 23607096 Free PMC article. Review.
-
Stabilizing Off-pathway Oligomers by Polyphenol Nanoassemblies for IAPP Aggregation Inhibition.Sci Rep. 2016 Jan 14;6:19463. doi: 10.1038/srep19463. Sci Rep. 2016. PMID: 26763863 Free PMC article.
-
Implications of peptide assemblies in amyloid diseases.Chem Soc Rev. 2017 Oct 30;46(21):6492-6531. doi: 10.1039/c7cs00372b. Chem Soc Rev. 2017. PMID: 28702523 Free PMC article. Review.
-
Advanced protein formulations.Protein Sci. 2015 Jul;24(7):1031-9. doi: 10.1002/pro.2684. Epub 2015 May 1. Protein Sci. 2015. PMID: 25858529 Free PMC article. Review.
References
-
- Kopito R.R., Ron D. Conformational disease. Nat. Cell Biol. 2000;2:E207–E209. - PubMed
-
- Porat Y., Abramowitz A., Gazit E. Inhibition of amyloid fibril formation by polyphenols: structural similarity and aromatic interactions as a common inhibition mechanism. Chem. Biol. Drug Des. 2006;67:27–37. - PubMed
-
- Haass C., Selkoe D.J. Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer's amyloid β-peptide. Nat. Rev. Mol. Cell Biol. 2007;8:101–112. - PubMed
-
- Bucciantini M., Giannoni E., Stefani M. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature. 2002;416:507–511. - PubMed
-
- Kayed R., Head E., Glabe C.G. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 2003;300:486–489. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

