Extracellular DNA-dependent biofilm formation by Staphylococcus epidermidis RP62A in response to subminimal inhibitory concentrations of antibiotics
- PMID: 21402153
- PMCID: PMC3109171
- DOI: 10.1016/j.resmic.2011.03.008
Extracellular DNA-dependent biofilm formation by Staphylococcus epidermidis RP62A in response to subminimal inhibitory concentrations of antibiotics
Abstract
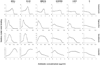
We measured the ability of Staphylococcus epidermidis to form biofilms in the presence of subminimal inhibitory concentrations (sub-MICs) of vancomycin, tigecycline, linezolid and novobiocin. Six strains that produce different amounts of biofilm were tested. The three strains that produced the highest amounts of biofilm exhibited steady-state or decreased biofilm formation in the presence of sub-MIC antibiotics, whereas the three strains that produced lower amounts of biofilm exhibited up to 10-fold-increased biofilm formation in the presence of sub-MIC antibiotics. In two of the inducible strains (9142 and 456a), antibiotic-induced biofilm formation was inhibited by dispersin B, an enzyme that degrades poly-N-acetylglucosamine (PNAG) biofilm polysaccharide. In the third inducible strain (RP62A), dispersin B inhibited biofilm formation in response to sub-MIC vancomycin, but not to sub-MIC tigecycline. In contrast, DNase I efficiently inhibited biofilm formation by strain RP62A in response to sub-MIC tigecycline and vancomycin. DNase I had no effect on antibiotic-induced biofilm formation in strains 9142 and 456a. Our findings indicate that antibiotic-induced biofilm formation in S. epidermidis is both strain- and antibiotic-dependent and that S. epidermidis RP62A utilizes an extracellular DNA-dependent mechanism to form biofilms in response to sub-MIC antibiotics.
Copyright © 2011 Institut Pasteur. Published by Elsevier SAS. All rights reserved.
Figures


References
-
- Boehm A, Steiner S, Zaehringer F, Casanova A, Hamburger F, Ritz D, Keck W, Ackermann M, Schirmer T, Jenal U. Second messenger signalling governs Escherichia coli biofilm induction upon ribosomal stress. Mol. Microbiol. 2009;72:1500–1516. - PubMed
-
- Cargill JS, Upton M. Low concentrations of vancomycin stimulate biofilm formation in some clinical isolates of Staphylococcus epidermidis. J. Clin. Pathol. 2009;62:1112–1116. - PubMed
-
- Chokr A, Watier D, Eleaume H, Pangon B, Ghnassia J-C, Mack D, Jabbouri S. Correlation between biofilm formation and production of polysaccharide intercellular adhesin in clinical isolates of coagulase-negative staphylococci. Int. J. Med. Microbiol. 2006;296:381–388. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

