Purification of histone ubiquitin ligases from HeLa cells
- PMID: 21402158
- PMCID: PMC3119374
- DOI: 10.1016/j.ymeth.2011.03.003
Purification of histone ubiquitin ligases from HeLa cells
Abstract
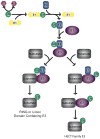
Posttranslational histone modifications play an important role in regulating chromatin based nuclear processes including transcription. Of these modifications, histone ubiquitination is among the least understood. Histone ubiquitination predominately targets histones H2A and H2B. While ubiquitination of H2B is evolutionarily conserved from budding yeast to mammals, ubiquitination of H2A has not been detected in budding yeast, worms, or plants. Until recently, studies of histone ubiquitination lagged far behind the study of other histone modifications, largely because antibodies specific for ubiquitinated histones are difficult to generate. Despite this obstacle, the identification of the enzymatic machineries involved in histone ubiquitination, together with the successful use of a combination of genetic and immunoblot approaches to detect ubiquitinated histones, have helped to reveal important regulatory roles for this modification in transcriptional initiation and elongation, cell cycle progression, and DNA damage response. With the aid of the recently developed ubiquitinated histone-specific antibodies, an intriguing link between histone ubiquitination and cancer development has been established. While the enzymes involved in H2B ubiquitination were identified first in budding yeast and subsequently in higher organisms based on gene homology, the identification of the enzymatic machineries involved in H2A ubiquitination largely depended on a biochemical purification approach. The unbiased search for ubiquitin ligases targeting histones also led to the identification of a H3 and H4 ubiquitin ligase. Here we detail a protocol for the biochemical approach to identify histone ubiquitin ligase(s) from HeLa cells. Similar approaches have been successfully used to identify histone methyltransferases, histone demethylases, chromatin remodeling factors, and general transcription factors. So long as an in vitro enzymatic assay can be established, the approach we describe can be easily adapted to identify other histone and non-histone modifying enzymes.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures




Similar articles
-
In Vitro Assay to Study Histone Ubiquitination During Transcriptional Regulation.Methods Mol Biol. 2017;1507:235-244. doi: 10.1007/978-1-4939-6518-2_17. Methods Mol Biol. 2017. PMID: 27832544
-
Role of histone H2A ubiquitination in Polycomb silencing.Nature. 2004 Oct 14;431(7010):873-8. doi: 10.1038/nature02985. Epub 2004 Sep 22. Nature. 2004. PMID: 15386022
-
In vitro and in vivo assays for studying histone ubiquitination and deubiquitination.Methods Mol Biol. 2009;523:295-309. doi: 10.1007/978-1-59745-190-1_20. Methods Mol Biol. 2009. PMID: 19381930
-
Histone ubiquitination: triggering gene activity.Mol Cell. 2008 Mar 28;29(6):653-63. doi: 10.1016/j.molcel.2008.02.014. Mol Cell. 2008. PMID: 18374642 Review.
-
The elusive structural role of ubiquitinated histones.Biochem Cell Biol. 2002;80(3):311-9. doi: 10.1139/o02-081. Biochem Cell Biol. 2002. PMID: 12123284 Review.
Cited by
-
Role of remodeling and spacing factor 1 in histone H2A ubiquitination-mediated gene silencing.Proc Natl Acad Sci U S A. 2017 Sep 19;114(38):E7949-E7958. doi: 10.1073/pnas.1711158114. Epub 2017 Aug 30. Proc Natl Acad Sci U S A. 2017. PMID: 28855339 Free PMC article.
-
The histone H2A deubiquitinase USP16 interacts with HERC2 and fine-tunes cellular response to DNA damage.J Biol Chem. 2014 Nov 21;289(47):32883-94. doi: 10.1074/jbc.M114.599605. Epub 2014 Oct 10. J Biol Chem. 2014. PMID: 25305019 Free PMC article.
-
Divergent Mechanisms of H2AZ.1 and H2AZ.2 in PRC1-Mediated H2A Ubiquitination.Cells. 2025 Jul 23;14(15):1133. doi: 10.3390/cells14151133. Cells. 2025. PMID: 40801566 Free PMC article.
-
Epigenetic mechanisms in developmental programming of adult disease.Drug Discov Today. 2011 Dec;16(23-24):1007-18. doi: 10.1016/j.drudis.2011.09.008. Epub 2011 Sep 16. Drug Discov Today. 2011. PMID: 21945859 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

