Importin beta plays an essential role in the regulation of the LysRS-Ap(4)A pathway in immunologically activated mast cells
- PMID: 21402779
- PMCID: PMC3133347
- DOI: 10.1128/MCB.01159-10
Importin beta plays an essential role in the regulation of the LysRS-Ap(4)A pathway in immunologically activated mast cells
Abstract
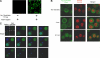
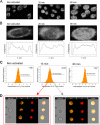
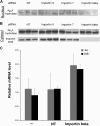
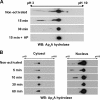
We recently reported that diadenosine tetraphosphate hydrolase (Ap(4)A hydrolase) plays a critical role in gene expression via regulation of intracellular Ap(4)A levels. This enzyme serves as a component of our newly described lysyl tRNA synthetase (LysRS)-Ap(4)A biochemical pathway that is triggered upon immunological challenge. Here we explored the mechanism of this enzyme's translocation into the nucleus and found its immunologically dependent association with importin beta. Silencing of importin beta prevented Ap(4)A hydrolase nuclear translocation and affected the local concentration of Ap(4)A, which led to an increase in microphthalmia transcription factor (MITF) transcriptional activity. Furthermore, immunological activation of mast cells resulted in dephosphorylation of Ap(4)A hydrolase, which changed the hydrolytic activity of the enzyme.
Figures



References
-
- Abdelghany H. M., et al. 2001. Cloning, characterisation and crystallisation of a diadenosine 5′, 5‴-P(1),P(4)-tetraphosphate pyrophosphohydrolase from Caenorhabditis elegans. Biochim. Biophys. Acta 1550:27–36 - PubMed
-
- Bayliss R., Littlewood T., Strawn L. A., Wente S. R., Stewart M. 2002. GLFG and FxFG nucleoporins bind to overlapping sites on importin-beta. J. Biol. Chem. 277:50597–50606 - PubMed
-
- Blom N., Gammeltoft S., Brunak S. 1999. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 294:1351–1362 - PubMed
-
- Reference deleted.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
