Stanniocalcin has deep evolutionary roots in eukaryotes
- PMID: 21402861
- PMCID: PMC5654410
- DOI: 10.1093/gbe/evr020
Stanniocalcin has deep evolutionary roots in eukaryotes
Abstract
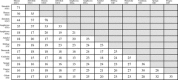
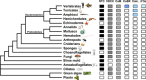
Vertebrates have a large glycoprotein hormone, stanniocalcin, which originally was shown to inhibit calcium uptake from the environment in teleost fish gills. Later, humans, other mammals, and teleost fish were shown to have two forms of stanniocalcin (STC1 and STC2) that were widely distributed in many tissues. STC1 is associated with calcium and phosphate homeostasis and STC2 with phosphate, but their receptors and signaling pathways have not been elucidated. We undertook a phylogenetic investigation of stanniocalcin beyond the vertebrates using a combination of BLAST and HMMER homology searches in protein, genomic, and expressed sequence tag databases. We identified novel STC homologs in a diverse array of multicellular and unicellular organisms. Within the eukaryotes, almost all major taxonomic groups except plants and algae have STC homologs, although some groups like echinoderms and arthropods lack STC genes. The critical structural feature for recognition of stanniocalcins was the conserved pattern of ten cysteines, even though the amino acid sequence identity was low. Signal peptides in STC sequences suggest they are secreted from the cell of synthesis. The role of glycosylation signals and additional cysteines is not yet clear, although the 11th cysteine, if present, has been shown to form homodimers in some vertebrates. We predict that large secreted stanniocalcin homologs appeared in evolution as early as single-celled eukaryotes. Stanniocalcin's tertiary structure with five disulfide bonds and its primary structure with modest amino acid conservation currently lack an established receptor-signaling system, although we suggest possible alternatives.
Figures


References
-
- Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. - PubMed
-
- Albrand JP, Blackledge MJ, Pascaud F, Hollecker M, Marion D. NMR and restrained molecular dynamics study of the three-dimensional solution structure of toxin FS2, a specific blocker of the L-type calcium channel, isolated from black mamba venom. Biochemistry. 1995;34:5923–5937. - PubMed
-
- Amemiya Y, Marra LE, Reyhani N, Youson JH. Stanniocalcin from an ancient teleost: a monomeric form of the hormone and a possible extracorpuscular distribution. Mol Cell Endocrinol. 2002;188:141–150. - PubMed
-
- Amemiya Y, Youson JH. Primary structure of stanniocalcin in two basal Actinopterygii. Gen Comp Endocrinol. 2004;135:250–257. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

