Structural biology of Gram-positive bacterial adhesins
- PMID: 21404359
- PMCID: PMC3125861
- DOI: 10.1002/pro.613
Structural biology of Gram-positive bacterial adhesins
Abstract
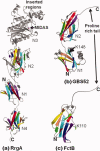
The structural biology of Gram-positive cell surface adhesins is an emerging field of research, whereas Gram-negative pilus assembly and anchoring have been extensively investigated and are well understood. Gram-positive surface proteins known as MSCRAMMs (microbial surface components recognizing adhesive matrix molecules) and individual proteins that assemble into long, hair-like organelles known as pili have similar features at the primary sequence level as well as at the tertiary structural level. Some of these conserved features are essential for their transportation from the cytoplasm and for cell wall anchoring. More importantly, the MSCRAMMs and the individual pilins are assembled with building blocks that are variants of structural modules used for human immunoglobulins. MSCRAMMs target the host's extracellular matrix proteins, such as collagen, fibrinogen, and fibronectin, and they have received considerable attention from structural biologists in the last decade, who have primarily been interested in understanding their interactions with host tissue. The recent focus is on the newly discovered pili of Gram-positive bacteria, and in this review, we highlight the advances in understanding of the individual pilus constituents and their associations and stress the similarities between the individual pilins and surface proteins.
Copyright © 2011 The Protein Society.
Figures


Similar articles
-
Structure, Function, and Assembly of Adhesive Organelles by Uropathogenic Bacteria.Microbiol Spectr. 2015 Oct;3(5):10.1128/microbiolspec.UTI-0018-2013. doi: 10.1128/microbiolspec.UTI-0018-2013. Microbiol Spectr. 2015. PMID: 26542038 Free PMC article. Review.
-
Crystallography of gram-positive bacterial adhesins.Adv Exp Med Biol. 2011;715:175-95. doi: 10.1007/978-94-007-0940-9_11. Adv Exp Med Biol. 2011. PMID: 21557064 Review.
-
MSCRAMM-mediated adherence of microorganisms to host tissues.Annu Rev Microbiol. 1994;48:585-617. doi: 10.1146/annurev.mi.48.100194.003101. Annu Rev Microbiol. 1994. PMID: 7826020 Review.
-
Identification and phenotypic characterization of a second collagen adhesin, Scm, and genome-based identification and analysis of 13 other predicted MSCRAMMs, including four distinct pilus loci, in Enterococcus faecium.Microbiology (Reading). 2008 Oct;154(Pt 10):3199-3211. doi: 10.1099/mic.0.2008/017319-0. Microbiology (Reading). 2008. PMID: 18832325 Free PMC article.
-
Pilins in gram-positive bacteria: A structural perspective.IUBMB Life. 2015 Jul;67(7):533-43. doi: 10.1002/iub.1400. Epub 2015 Jul 14. IUBMB Life. 2015. PMID: 26178080 Review.
Cited by
-
Structure of Streptococcus agalactiae tip pilin GBS104: a model for GBS pili assembly and host interactions.Acta Crystallogr D Biol Crystallogr. 2013 Jun;69(Pt 6):1073-89. doi: 10.1107/S0907444913004642. Epub 2013 May 15. Acta Crystallogr D Biol Crystallogr. 2013. PMID: 23695252 Free PMC article.
-
Mycobacterium tuberculosis Primary Infection and Dissemination: A Critical Role for Alveolar Epithelial Cells.Front Cell Infect Microbiol. 2019 Aug 21;9:299. doi: 10.3389/fcimb.2019.00299. eCollection 2019. Front Cell Infect Microbiol. 2019. PMID: 31497538 Free PMC article. Review.
-
LrpCBA pilus proteins of gut-dwelling Ligilactobacillus ruminis: crystallization and X-ray diffraction analysis.Acta Crystallogr F Struct Biol Commun. 2021 Aug 1;77(Pt 8):238-245. doi: 10.1107/S2053230X21007263. Epub 2021 Jul 28. Acta Crystallogr F Struct Biol Commun. 2021. PMID: 34341189 Free PMC article.
-
Structure, Function, and Assembly of Adhesive Organelles by Uropathogenic Bacteria.Microbiol Spectr. 2015 Oct;3(5):10.1128/microbiolspec.UTI-0018-2013. doi: 10.1128/microbiolspec.UTI-0018-2013. Microbiol Spectr. 2015. PMID: 26542038 Free PMC article. Review.
-
Bioadhesion in the oral cavity and approaches for biofilm management by surface modifications.Clin Oral Investig. 2020 Dec;24(12):4237-4260. doi: 10.1007/s00784-020-03646-1. Epub 2020 Oct 27. Clin Oral Investig. 2020. PMID: 33111157 Free PMC article. Review.
References
-
- Sauer FG, Mulvey MA, Schilling JD, Martinez JJ, Hultgren SJ. Bacterial pili: molecular mechanisms of pathogenesis. Curr Opin Microbiol. 2000;3:65–72. - PubMed
-
- Choudhury D, Thompson A, Stojanoff V, Langermann S, Pinkner J, Hultgren SJ, Knight SD. X-ray structure of the FimC-FimH chaperone-adhesin complex from uropathogenic Escherichia coli. Science. 1999;285:1061–1066. - PubMed
-
- Scott JR, Zahner D. Pili with strong attachments: Gram-positive bacteria do it differently. Mol Microbiol. 2006;62:320–330. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

