Protein sliding and hopping kinetics on DNA
- PMID: 21405863
- PMCID: PMC3683889
- DOI: 10.1103/PhysRevE.83.021907
Protein sliding and hopping kinetics on DNA
Abstract
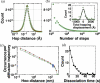
Using Monte Carlo simulations, we deconvolved the sliding and hopping kinetics of GFP-LacI proteins on elongated DNA from their experimentally observed seconds-long diffusion trajectories. Our simulations suggest the following results: (i) in each diffusion trajectory, a protein makes on average hundreds of alternating slides and hops with a mean sliding time of several tens of milliseconds; (ii) sliding dominates the root-mean-square displacement of fast diffusion trajectories, whereas hopping dominates slow ones; (iii) flow and variations in salt concentration have limited effects on hopping kinetics, while in vivo DNA configuration is not expected to influence sliding kinetics; and (iv) the rate of occurrence for hops longer than 200 nm agrees with experimental data for EcoRV proteins.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
