COLD-PCR: improving the sensitivity of molecular diagnostics assays
- PMID: 21405967
- PMCID: PMC3111913
- DOI: 10.1586/erm.10.115
COLD-PCR: improving the sensitivity of molecular diagnostics assays
Abstract
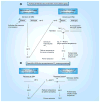
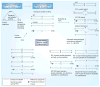
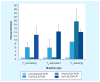
The detection of low-abundance DNA variants or mutations is of particular interest to medical diagnostics, individualized patient treatment and cancer prognosis; however, detection sensitivity for low-abundance variants is a pronounced limitation of most currently available molecular assays. We have recently developed coamplification at lower denaturation temperature-PCR (COLD-PCR) to resolve this limitation. This novel form of PCR selectively amplifies low-abundance DNA variants from mixtures of wild-type and mutant-containing (or variant-containing) sequences, irrespective of the mutation type or position on the amplicon, by using a critical denaturation temperature. The use of a lower denaturation temperature in COLD-PCR results in selective denaturation of amplicons with mutation-containing molecules within wild-type mutant heteroduplexes or with a lower melting temperature. COLD-PCR can be used in lieu of conventional PCR in several molecular applications, thus enriching the mutant fraction and improving the sensitivity of downstream mutation detection by up to 100-fold.
Figures



References
Websites
-
- MeltSim: The DNA Melting Simulator. www.bioinformatics.org/meltsim/wiki.
-
- Institut für Physikalische Biologie. www.biophys.uni-duesseldorf.de/local/POLAND/poland.html.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
