Analog regulation of metabolic demand
- PMID: 21406074
- PMCID: PMC3068955
- DOI: 10.1186/1752-0509-5-40
Analog regulation of metabolic demand
Abstract
Background: The 3D structure of the chromosome of the model organism Escherichia coli is one key component of its gene regulatory machinery. This type of regulation mediated by topological transitions of the chromosomal DNA can be thought of as an analog control, complementing the digital control, i.e. the network of regulation mediated by dedicated transcription factors. It is known that alterations in the superhelical density of chromosomal DNA lead to a rich pattern of differential expressed genes. Using a network approach, we analyze these expression changes for wild type E. coli and mutants lacking nucleoid associated proteins (NAPs) from a metabolic and transcriptional regulatory network perspective.
Results: We find a significantly higher correspondence between gene expression and metabolism for the wild type expression changes compared to mutants in NAPs, indicating that supercoiling induces meaningful metabolic adjustments. As soon as the underlying regulatory machinery is impeded (as for the NAP mutants), this coherence between expression changes and the metabolic network is substantially reduced. This effect is even more pronounced, when we compute a wild type metabolic flux distribution using flux balance analysis and restrict our analysis to active reactions. Furthermore, we are able to show that the regulatory control exhibited by DNA supercoiling is not mediated by the transcriptional regulatory network (TRN), as the consistency of the expression changes with the TRN logic of activation and suppression is strongly reduced in the wild type in comparison to the mutants.
Conclusions: So far, the rich patterns of gene expression changes induced by alterations of the superhelical density of chromosomal DNA have been difficult to interpret. Here we characterize the effective networks formed by supercoiling-induced gene expression changes mapped onto reconstructions of E. coli's metabolic and transcriptional regulatory network. Our results show that DNA supercoiling coordinates gene expression with metabolism. Furthermore, this control is acting directly because we can exclude the potential role of the TRN as a mediator.
Figures




 , (6) iAF1260man obtained from FBA (rich medium), (7) iAF1260man, (8) EcoCyc**, (9) KEGG*, (10) iAF1260*, (11) iAF1260deg, (12)
, (6) iAF1260man obtained from FBA (rich medium), (7) iAF1260man, (8) EcoCyc**, (9) KEGG*, (10) iAF1260*, (11) iAF1260deg, (12)  , (13) Flux-coupling network (fully coupled), (14) KEGG, (15) Intersection of KEGG and iAF1260man, (16) Intersection of EcoCyc, KEGG and iAF1260man, (17) KEGG**, (18) Flux-coupling network (fully and directionally coupled), (19)
, (13) Flux-coupling network (fully coupled), (14) KEGG, (15) Intersection of KEGG and iAF1260man, (16) Intersection of EcoCyc, KEGG and iAF1260man, (17) KEGG**, (18) Flux-coupling network (fully and directionally coupled), (19)  , (20) iAF1260**, (21) Flux-coupling network (directionally coupled), (22) iAF1260, (23) iAF1260*.
, (20) iAF1260**, (21) Flux-coupling network (directionally coupled), (22) iAF1260, (23) iAF1260*.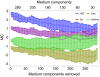

References
-
- Muskhelishvili G, Travers A. In: RNA polymerases as molecular motors. Buc H, Strick T, editor. Cambridge, UK: RSC Publishing; 2009. Intrinsic in vivo modulators: negative supercoiling and the constituents of the bacterial nucleoid; pp. 69–95. full_text.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

