CARMA3 is crucial for EGFR-Induced activation of NF-κB and tumor progression
- PMID: 21406399
- PMCID: PMC3059846
- DOI: 10.1158/0008-5472.CAN-10-3626
CARMA3 is crucial for EGFR-Induced activation of NF-κB and tumor progression
Abstract
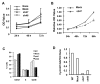
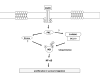
EGF activates NF-κB, and constitutively activated NF-κB contributes to EGFR mutation-associated tumorigenesis, but it remains unclear precisely how EGFR signaling leads to NF-κB activation. Here we report that CARMA3, a caspase recruitment domain (CARD)-containing scaffold molecule, is required for EGF-induced NF-κB activation. CARMA3 deficiency impaired the activation of the IKK complex following EGF stimulation, resulting in a defect of EGF-induced IκBα phosphorylation and NF-κB activation. We found that CARMA3 and Bcl10 contributed to several characteristics of EGFR-associated malignancy, including proliferation, survival, migration, and invasion. Most importantly, CARMA3 contributed to tumor growth in vivo. Our findings elucidate a crucial link between EGFR-proximal signaling components and the downstream IKK complex, and they suggest a new therapeutic target for treatment of EGFR-driven cancers.
©2011 AACR.
Figures




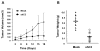
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

