Efficient gene transfer in bacterial cell chains
- PMID: 21406598
- PMCID: PMC3055163
- DOI: 10.1128/mBio.00027-11
Efficient gene transfer in bacterial cell chains
Abstract
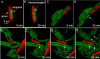
Horizontal gene transfer contributes to evolution and the acquisition of new traits. In bacteria, horizontal gene transfer is often mediated by conjugative genetic elements that transfer directly from cell to cell. Integrative and conjugative elements (ICEs; also known as conjugative transposons) are mobile genetic elements that reside within a host genome but can excise to form a circle and transfer by conjugation to recipient cells. ICEs contribute to the spread of genes involved in pathogenesis, symbiosis, metabolism, and antibiotic resistance. Despite its importance, little is known about the mechanisms of conjugation in Gram-positive bacteria or how quickly or frequently transconjugants become donors. We visualized the transfer of the integrative and conjugative element ICEBs1 from a Bacillus subtilis donor to recipient cells in real time using fluorescence microscopy. We found that transfer of DNA from a donor to a recipient appeared to occur at a cell pole or along the lateral cell surface of either cell. Most importantly, we found that when acquired by 1 cell in a chain, ICEBs1 spread rapidly from cell to cell within the chain by additional sequential conjugation events. This intrachain conjugation is inherently more efficient than conjugation that is due to chance encounters between individual cells. Many bacterial species, including pathogenic, commensal, symbiotic, and nitrogen-fixing organisms, harbor ICEs and grow in chains, often as parts of microbial communities. It is likely that efficient intrachain spreading is a general feature of conjugative DNA transfer and serves to amplify the number of cells that acquire conjugative mobile genetic elements. IMPORTANCE Conjugative elements contribute to horizontal gene transfer and the acquisition of new traits. They are largely responsible for spreading antibiotic resistance in bacterial communities. To study the cell biology of conjugation, we visualized conjugative DNA transfer between Bacillus subtilis cells in real time using fluorescence microscopy. In contrast to previous predictions that transfer would occur preferentially from the donor cell pole, we found that transfer of DNA from a donor to a recipient appeared to occur at a cell pole or along the lateral cell surface of either cell. Most importantly, we found that when acquired by 1 cell in a chain, the conjugative DNA spread rapidly from cell to cell within the chain through sequential conjugation events. Since many bacterial species grow naturally in chains, this intrachain transfer is likely a common mechanism for accelerating the spread of conjugative elements within microbial communities.
Figures
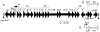

Similar articles
-
Specificity and Selective Advantage of an Exclusion System in the Integrative and Conjugative Element ICEBs1 of Bacillus subtilis.J Bacteriol. 2021 Apr 21;203(10):e00700-20. doi: 10.1128/JB.00700-20. Print 2021 Apr 21. J Bacteriol. 2021. PMID: 33649151 Free PMC article.
-
Biofilm Formation Drives Transfer of the Conjugative Element ICEBs1 in Bacillus subtilis.mSphere. 2018 Sep 26;3(5):e00473-18. doi: 10.1128/mSphere.00473-18. mSphere. 2018. PMID: 30258041 Free PMC article.
-
Biology of ICEBs1, an integrative and conjugative element in Bacillus subtilis.Plasmid. 2016 Jul;86:14-25. doi: 10.1016/j.plasmid.2016.07.001. Epub 2016 Jul 2. Plasmid. 2016. PMID: 27381852 Review.
-
Characterization of ConE, the VirB4 Homolog of the Integrative and Conjugative Element ICEBs1 of Bacillus subtilis.J Bacteriol. 2023 Jun 27;205(6):e0003323. doi: 10.1128/jb.00033-23. Epub 2023 May 23. J Bacteriol. 2023. PMID: 37219457 Free PMC article.
-
Integrative and Conjugative Elements (ICEs): What They Do and How They Work.Annu Rev Genet. 2015;49:577-601. doi: 10.1146/annurev-genet-112414-055018. Epub 2015 Oct 14. Annu Rev Genet. 2015. PMID: 26473380 Free PMC article. Review.
Cited by
-
Genome plasticity and systems evolution in Streptomyces.BMC Bioinformatics. 2012 Jun 25;13 Suppl 10(Suppl 10):S8. doi: 10.1186/1471-2105-13-S10-S8. BMC Bioinformatics. 2012. PMID: 22759432 Free PMC article.
-
Monitoring Bacterial Conjugation by Optical Microscopy.Front Microbiol. 2021 Oct 4;12:750200. doi: 10.3389/fmicb.2021.750200. eCollection 2021. Front Microbiol. 2021. PMID: 34671336 Free PMC article.
-
Specificity and Selective Advantage of an Exclusion System in the Integrative and Conjugative Element ICEBs1 of Bacillus subtilis.J Bacteriol. 2021 Apr 21;203(10):e00700-20. doi: 10.1128/JB.00700-20. Print 2021 Apr 21. J Bacteriol. 2021. PMID: 33649151 Free PMC article.
-
Critical Components of the Conjugation Machinery of the Integrative and Conjugative Element ICEBs1 of Bacillus subtilis.J Bacteriol. 2015 Aug 1;197(15):2558-67. doi: 10.1128/JB.00142-15. Epub 2015 May 26. J Bacteriol. 2015. PMID: 26013486 Free PMC article.
-
Unveiling intraspecific diversity and evolutionary dynamics of the foodborne pathogen Bacillus paranthracis through high-quality pan-genome analysis.Curr Res Food Sci. 2024 Sep 21;9:100867. doi: 10.1016/j.crfs.2024.100867. eCollection 2024. Curr Res Food Sci. 2024. PMID: 39376581 Free PMC article.
References
-
- Ochman H, Lawrence JG, Groisman EA. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299–304 - PubMed
-
- Frost LS, Leplae R, Summers AO, Toussaint A. 2005. Mobile genetic elements: the agents of open source evolution. Nat. Rev. Microbiol. 3:722–732 - PubMed
-
- Thomas CM, Nielsen KM. 2005. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 3:711–721 - PubMed
-
- Wozniak RA, Waldor MK. 2010. Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat. Rev. Microbiol. 8:552–563 - PubMed
-
- Burrus V, Pavlovic G, Decaris B, Guedon G. 2002. Conjugative transposons: the tip of the iceberg. Mol. Microbiol. 46:601–610 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

